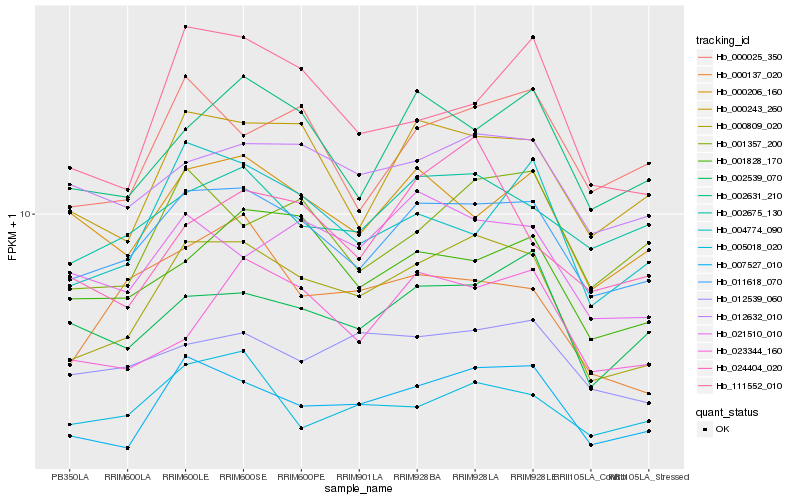
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000809_020 |
0.0 |
desease resistance |
Gene Name: RPW8 |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_007527_010 |
0.0806994827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_011618_070 |
0.0899182041 |
- |
- |
NMDA receptor-regulated protein [Medicago truncatula] |
| 4 |
Hb_000243_260 |
0.1178453147 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 5 |
Hb_111552_010 |
0.1245075243 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 6 |
Hb_012539_060 |
0.1265462888 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 7 |
Hb_012632_010 |
0.1274100202 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 8 |
Hb_004774_090 |
0.1276162489 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 9 |
Hb_021510_010 |
0.1281578381 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 10 |
Hb_000137_020 |
0.1290630644 |
- |
- |
PREDICTED: ras-related protein RABA5b [Jatropha curcas] |
| 11 |
Hb_001357_200 |
0.1310970891 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_001828_170 |
0.1313849277 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 13 |
Hb_002631_210 |
0.1317504805 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 14 |
Hb_002539_070 |
0.1318139322 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 15 |
Hb_023344_160 |
0.133739404 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 16 |
Hb_005018_020 |
0.1349726634 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g07740, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000206_160 |
0.1365702778 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_002675_130 |
0.1370062711 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 16, NatA auxiliary subunit isoform X1 [Jatropha curcas] |
| 19 |
Hb_024404_020 |
0.1374830831 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 20 |
Hb_000025_350 |
0.1377259673 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |