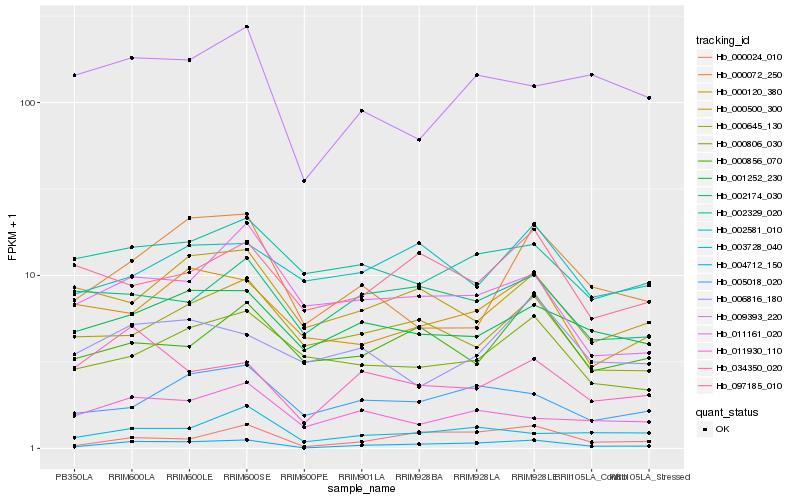
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004712_150 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000024_010 |
0.1401344849 |
- |
- |
Uncharacterized protein TCM_044274 [Theobroma cacao] |
| 3 |
Hb_005018_020 |
0.1831705368 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g07740, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001252_230 |
0.1907612065 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003728_040 |
0.1908748389 |
- |
- |
PREDICTED: uncharacterized protein LOC101492305 [Cicer arietinum] |
| 6 |
Hb_000806_030 |
0.1926050534 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 7 |
Hb_000645_130 |
0.1933242757 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_000120_380 |
0.1940326445 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 9 |
Hb_000856_070 |
0.1959514237 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 10 |
Hb_000072_250 |
0.1976624626 |
transcription factor |
TF Family: Orphans |
- |
| 11 |
Hb_002174_030 |
0.1977193262 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 12 |
Hb_011161_020 |
0.197963661 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 13 |
Hb_034350_020 |
0.1987473519 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 14 |
Hb_011930_110 |
0.2015790227 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 15 |
Hb_002329_020 |
0.204147668 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 16 |
Hb_097185_010 |
0.2050370554 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 17 |
Hb_000500_300 |
0.2052480599 |
- |
- |
PREDICTED: nucleolar protein 10 [Jatropha curcas] |
| 18 |
Hb_002581_010 |
0.2066112488 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 19 |
Hb_009393_220 |
0.2066837748 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 20 |
Hb_006816_180 |
0.2067228544 |
- |
- |
- |