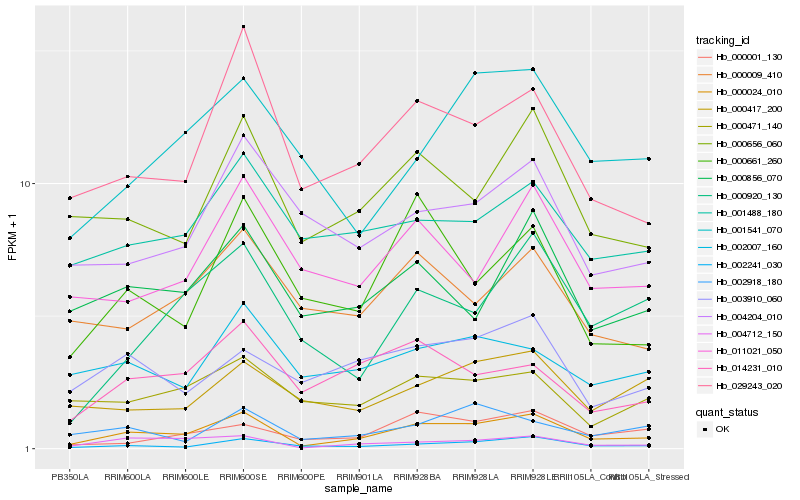
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000024_010 |
0.0 |
- |
- |
Uncharacterized protein TCM_044274 [Theobroma cacao] |
| 2 |
Hb_004712_150 |
0.1401344849 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_002241_030 |
0.1505691792 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 4 |
Hb_029243_020 |
0.1572507504 |
- |
- |
ENTH/VHS/GAT family protein [Theobroma cacao] |
| 5 |
Hb_000656_060 |
0.169525541 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_000661_260 |
0.1795679989 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 7 |
Hb_000920_130 |
0.1843977984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000856_070 |
0.187359333 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 9 |
Hb_014231_010 |
0.1876694527 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 10 |
Hb_000001_130 |
0.1889098701 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 11 |
Hb_001541_070 |
0.1914885942 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 12 |
Hb_011021_050 |
0.1924479224 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003910_060 |
0.1931043589 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g37170 [Jatropha curcas] |
| 14 |
Hb_002918_180 |
0.1937192253 |
- |
- |
hypothetical protein POPTR_0004s04740g [Populus trichocarpa] |
| 15 |
Hb_000009_410 |
0.1949964861 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 16 |
Hb_000471_140 |
0.1956861533 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g28640 [Prunus mume] |
| 17 |
Hb_000417_200 |
0.1978670362 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004204_010 |
0.1981580569 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 19 |
Hb_002007_160 |
0.1983864292 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 20 |
Hb_001488_180 |
0.1987096759 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |