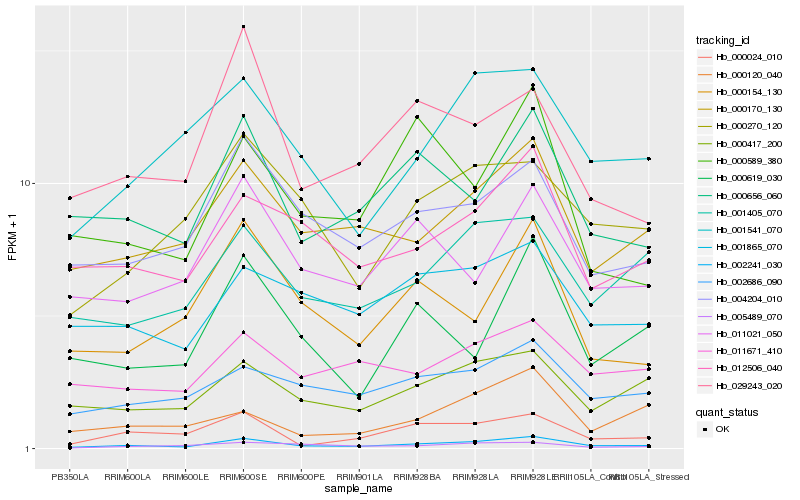
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002241_030 |
0.0 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 2 |
Hb_000024_010 |
0.1505691792 |
- |
- |
Uncharacterized protein TCM_044274 [Theobroma cacao] |
| 3 |
Hb_000417_200 |
0.1531748106 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000170_130 |
0.1547351392 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 5 |
Hb_000656_060 |
0.158463661 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_002686_090 |
0.1585436602 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 7 |
Hb_012506_040 |
0.1594399587 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 8 |
Hb_004204_010 |
0.1600804174 |
- |
- |
PREDICTED: uncharacterized protein LOC105640608 [Jatropha curcas] |
| 9 |
Hb_001541_070 |
0.1607267352 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1-like [Jatropha curcas] |
| 10 |
Hb_000154_130 |
0.1627006845 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 11 |
Hb_001405_070 |
0.1646776837 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 12 |
Hb_000619_030 |
0.1651969288 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_029243_020 |
0.1657633106 |
- |
- |
ENTH/VHS/GAT family protein [Theobroma cacao] |
| 14 |
Hb_000270_120 |
0.1669388723 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 15 |
Hb_005489_070 |
0.1707668939 |
- |
- |
PREDICTED: transposon Tf2-1 polyprotein isoform X1 [Zea mays] |
| 16 |
Hb_011021_050 |
0.1715540344 |
- |
- |
PREDICTED: uncharacterized protein LOC105644387 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000589_380 |
0.1720970114 |
- |
- |
Phosphoribosylformylglycinamidine synthase, putative [Ricinus communis] |
| 18 |
Hb_001865_070 |
0.1721587813 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011671_410 |
0.1727313174 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 20 |
Hb_000120_040 |
0.1744704857 |
- |
- |
- |