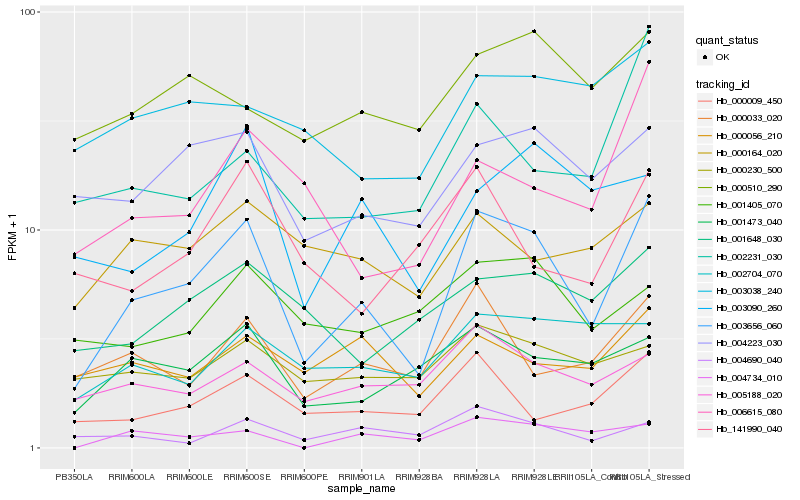
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003656_060 |
0.0 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 2 |
Hb_001473_040 |
0.1958955602 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_004223_030 |
0.1962907046 |
- |
- |
PREDICTED: uncharacterized protein LOC105635347 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000033_020 |
0.1971679982 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_005188_020 |
0.2023236818 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 6 |
Hb_000009_450 |
0.2034092208 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_003090_260 |
0.2073136651 |
- |
- |
hypothetical protein JCGZ_25097 [Jatropha curcas] |
| 8 |
Hb_004690_040 |
0.2110379823 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420 [Jatropha curcas] |
| 9 |
Hb_006615_080 |
0.2111465505 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001648_030 |
0.2116778551 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004734_010 |
0.2155577849 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002231_030 |
0.2158352439 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 13 |
Hb_002704_070 |
0.2158406886 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 14 |
Hb_000510_290 |
0.2164309499 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000056_210 |
0.2176846054 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26900, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001405_070 |
0.2191357611 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 17 |
Hb_141990_040 |
0.2196020215 |
- |
- |
Protein SCAR2, putative [Ricinus communis] |
| 18 |
Hb_000230_500 |
0.2202346072 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000164_020 |
0.2209506871 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 20 |
Hb_003038_240 |
0.2216995215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |