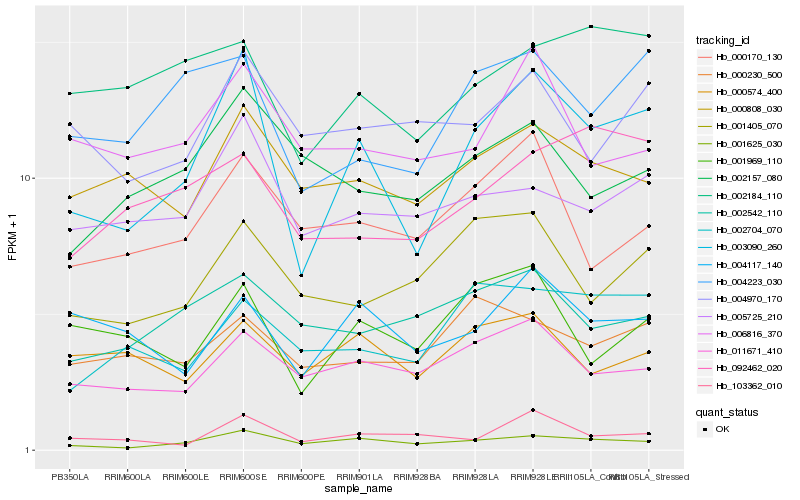
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003090_260 |
0.0 |
- |
- |
hypothetical protein JCGZ_25097 [Jatropha curcas] |
| 2 |
Hb_001625_030 |
0.1277929464 |
- |
- |
hypothetical protein JCGZ_20707 [Jatropha curcas] |
| 3 |
Hb_004223_030 |
0.1555171349 |
- |
- |
PREDICTED: uncharacterized protein LOC105635347 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011671_410 |
0.1647348516 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 5 |
Hb_005725_210 |
0.1656697529 |
- |
- |
PREDICTED: RNA-binding protein 1 [Jatropha curcas] |
| 6 |
Hb_001969_110 |
0.1685322824 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g15510, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002704_070 |
0.1714647191 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like [Jatropha curcas] |
| 8 |
Hb_000808_030 |
0.1728000502 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 9 |
Hb_004117_140 |
0.1732640089 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g16710, mitochondrial [Populus euphratica] |
| 10 |
Hb_002184_110 |
0.173752781 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 11 |
Hb_001405_070 |
0.174740195 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 12 |
Hb_103362_010 |
0.176259761 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_000170_130 |
0.1763467879 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 14 |
Hb_092462_020 |
0.1794656032 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000574_400 |
0.1814493225 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15930 [Jatropha curcas] |
| 16 |
Hb_004970_170 |
0.181714303 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 17 |
Hb_000230_500 |
0.1818960115 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 18 |
Hb_006816_370 |
0.1841072717 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 19 |
Hb_002157_080 |
0.1865067241 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002542_110 |
0.187418535 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |