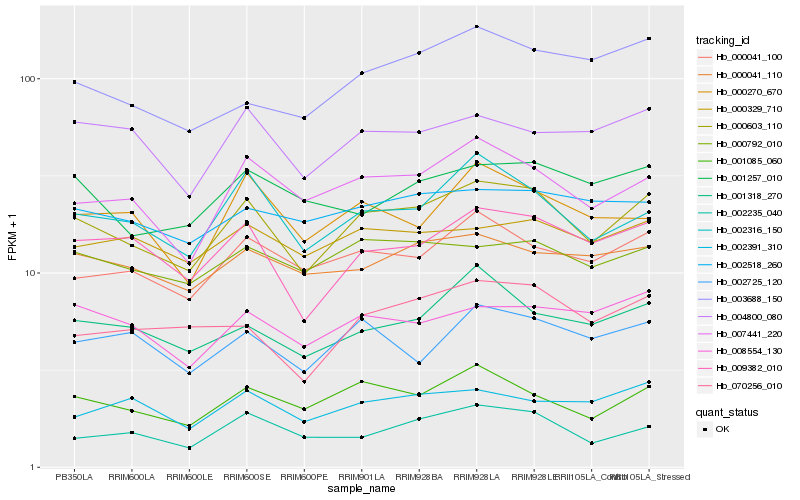
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000603_110 |
0.0 |
- |
- |
copper transporter, putative [Ricinus communis] |
| 2 |
Hb_009382_010 |
0.0756467263 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_001085_060 |
0.0800565551 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g04840 [Jatropha curcas] |
| 4 |
Hb_002316_150 |
0.0803944048 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 5 |
Hb_007441_220 |
0.0807016024 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 6 |
Hb_000041_100 |
0.0844650861 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 7 |
Hb_008554_130 |
0.0920641811 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105634317 [Jatropha curcas] |
| 8 |
Hb_070256_010 |
0.0921177443 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105630457 [Jatropha curcas] |
| 9 |
Hb_004800_080 |
0.0923910304 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 10 |
Hb_000792_010 |
0.0930525787 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 11 |
Hb_001318_270 |
0.0941029373 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 12 |
Hb_000041_110 |
0.0960353864 |
- |
- |
PREDICTED: WW domain-binding protein 11 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000329_710 |
0.0974985447 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001257_010 |
0.0980797094 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002725_120 |
0.0985567711 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002391_310 |
0.09965094 |
- |
- |
hypothetical protein PRUPE_ppa021315mg [Prunus persica] |
| 17 |
Hb_002518_260 |
0.1000488415 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 18 |
Hb_002235_040 |
0.1004579999 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 19 |
Hb_003688_150 |
0.1006883525 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000270_670 |
0.1012348923 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |