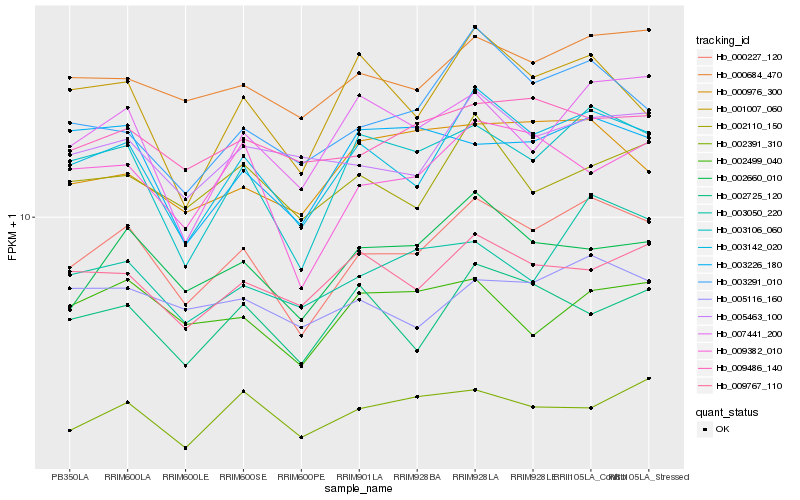
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003142_020 |
0.0524868866 |
- |
- |
PREDICTED: uncharacterized protein LOC105648334 [Jatropha curcas] |
| 3 |
Hb_003106_060 |
0.0656832597 |
- |
- |
hypothetical protein POPTR_0015s08140g [Populus trichocarpa] |
| 4 |
Hb_000684_470 |
0.0776838077 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 5 |
Hb_002660_010 |
0.0810428928 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18-like [Jatropha curcas] |
| 6 |
Hb_003291_010 |
0.0852002945 |
- |
- |
ATP-dependent protease La, putative [Ricinus communis] |
| 7 |
Hb_009382_010 |
0.0853411382 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_007441_200 |
0.0885110942 |
- |
- |
PREDICTED: uncharacterized protein LOC105643248 [Jatropha curcas] |
| 9 |
Hb_003050_220 |
0.0886987638 |
- |
- |
hypothetical protein JCGZ_04601 [Jatropha curcas] |
| 10 |
Hb_005463_100 |
0.0907766116 |
- |
- |
hypothetical protein POPTR_0001s08400g [Populus trichocarpa] |
| 11 |
Hb_009486_140 |
0.0929670755 |
- |
- |
PREDICTED: uncharacterized protein LOC105643248 [Jatropha curcas] |
| 12 |
Hb_005116_160 |
0.0933597162 |
transcription factor |
TF Family: TRAF |
- |
| 13 |
Hb_002110_150 |
0.0946258019 |
- |
- |
PREDICTED: serine/threonine-protein kinase PEPKR2 [Jatropha curcas] |
| 14 |
Hb_001007_060 |
0.0946663807 |
- |
- |
Transcriptional corepressor SEUSS [Glycine soja] |
| 15 |
Hb_009767_110 |
0.0953606842 |
- |
- |
- |
| 16 |
Hb_000976_300 |
0.0961488501 |
- |
- |
PREDICTED: uncharacterized protein LOC105646229 [Jatropha curcas] |
| 17 |
Hb_002499_040 |
0.0969126383 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase GCN2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002725_120 |
0.0969938024 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003226_180 |
0.0970427328 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 20 |
Hb_002391_310 |
0.0975201755 |
- |
- |
hypothetical protein PRUPE_ppa021315mg [Prunus persica] |