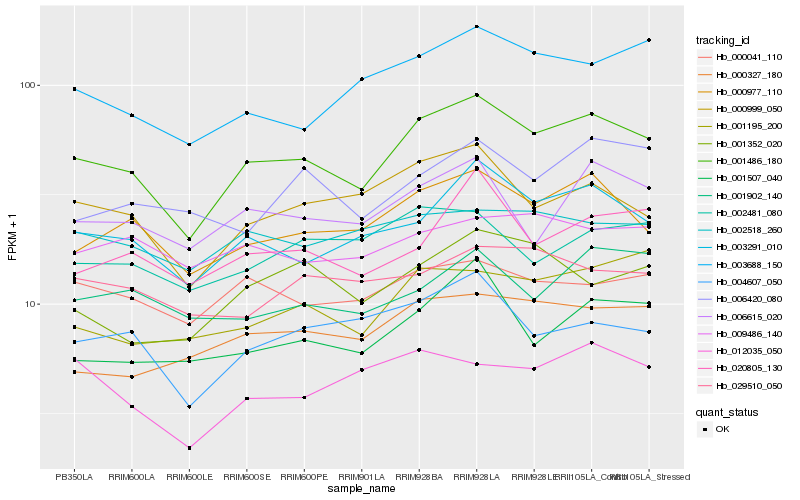
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001486_180 |
0.0 |
- |
- |
hypothetical protein B456_007G260200 [Gossypium raimondii] |
| 2 |
Hb_003291_010 |
0.0748213063 |
- |
- |
ATP-dependent protease La, putative [Ricinus communis] |
| 3 |
Hb_000977_110 |
0.0753570586 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000999_050 |
0.0853109873 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006615_020 |
0.0936857967 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 6 |
Hb_004607_050 |
0.0940986162 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_003688_150 |
0.0957447119 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 8 |
Hb_020805_130 |
0.0973312065 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 9 |
Hb_001507_040 |
0.0973954284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000041_110 |
0.0985474466 |
- |
- |
PREDICTED: WW domain-binding protein 11 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000327_180 |
0.1002895865 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001195_200 |
0.1007320357 |
- |
- |
PREDICTED: uncharacterized protein LOC105633802 [Jatropha curcas] |
| 13 |
Hb_002518_260 |
0.1028042054 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 14 |
Hb_002481_080 |
0.1029096611 |
- |
- |
PREDICTED: bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001902_140 |
0.1040747765 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 16 |
Hb_012035_050 |
0.1042861769 |
- |
- |
hypothetical protein JCGZ_10788 [Jatropha curcas] |
| 17 |
Hb_001352_020 |
0.1059552232 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_029510_050 |
0.1062158049 |
- |
- |
PREDICTED: golgin candidate 5 [Jatropha curcas] |
| 19 |
Hb_006420_080 |
0.1070983482 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_009486_140 |
0.1071994656 |
- |
- |
PREDICTED: uncharacterized protein LOC105643248 [Jatropha curcas] |