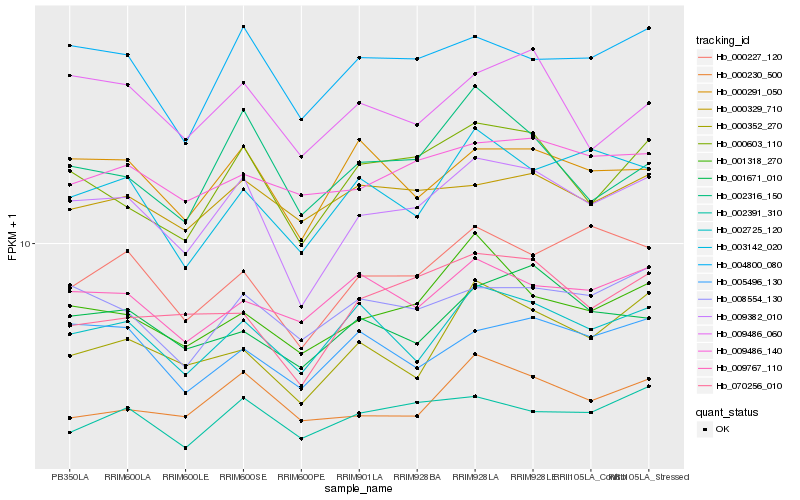
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009382_010 |
0.0 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_000603_110 |
0.0756467263 |
- |
- |
copper transporter, putative [Ricinus communis] |
| 3 |
Hb_004800_080 |
0.0778617391 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 4 |
Hb_002725_120 |
0.0832356298 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000227_120 |
0.0853411382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000291_050 |
0.0854092256 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008554_130 |
0.089252425 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105634317 [Jatropha curcas] |
| 8 |
Hb_001318_270 |
0.0896553837 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_070256_010 |
0.0922238644 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105630457 [Jatropha curcas] |
| 10 |
Hb_001671_010 |
0.0926078999 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 11 |
Hb_002391_310 |
0.092642517 |
- |
- |
hypothetical protein PRUPE_ppa021315mg [Prunus persica] |
| 12 |
Hb_005496_130 |
0.0926884706 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003142_020 |
0.0932157783 |
- |
- |
PREDICTED: uncharacterized protein LOC105648334 [Jatropha curcas] |
| 14 |
Hb_000230_500 |
0.0934420942 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 15 |
Hb_009486_060 |
0.0959396327 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 16 |
Hb_000352_270 |
0.0966528718 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26630, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_002316_150 |
0.0974995562 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 18 |
Hb_009486_140 |
0.0975974791 |
- |
- |
PREDICTED: uncharacterized protein LOC105643248 [Jatropha curcas] |
| 19 |
Hb_009767_110 |
0.0984160576 |
- |
- |
- |
| 20 |
Hb_000329_710 |
0.0989460903 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |