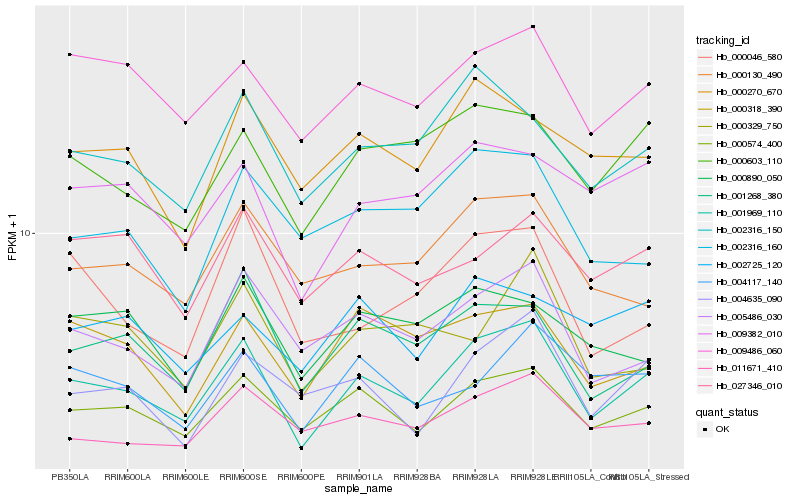
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001969_110 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g15510, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000574_400 |
0.0850916537 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15930 [Jatropha curcas] |
| 3 |
Hb_005486_030 |
0.1025471551 |
transcription factor |
TF Family: PHD |
hypothetical protein POPTR_0017s10890g [Populus trichocarpa] |
| 4 |
Hb_000603_110 |
0.103986713 |
- |
- |
copper transporter, putative [Ricinus communis] |
| 5 |
Hb_009486_060 |
0.1053292001 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 6 |
Hb_002316_150 |
0.1062092982 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 7 |
Hb_001268_380 |
0.1075063396 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g14080-like [Jatropha curcas] |
| 8 |
Hb_009382_010 |
0.107830098 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_011671_410 |
0.1121539095 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 10 |
Hb_000130_490 |
0.1139287082 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004635_090 |
0.1151504319 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 12 |
Hb_000318_390 |
0.115591777 |
- |
- |
PREDICTED: uncharacterized protein LOC105631693 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004117_140 |
0.1168990186 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g16710, mitochondrial [Populus euphratica] |
| 14 |
Hb_000270_670 |
0.1174615392 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 15 |
Hb_027346_010 |
0.1206019439 |
- |
- |
- |
| 16 |
Hb_000046_580 |
0.1213083476 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 17 |
Hb_002316_160 |
0.1240243365 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 18 |
Hb_000890_050 |
0.1249779128 |
- |
- |
PREDICTED: uncharacterized protein LOC105631800 [Jatropha curcas] |
| 19 |
Hb_000329_750 |
0.1254234581 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 20 |
Hb_002725_120 |
0.1256338961 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05750, chloroplastic [Jatropha curcas] |