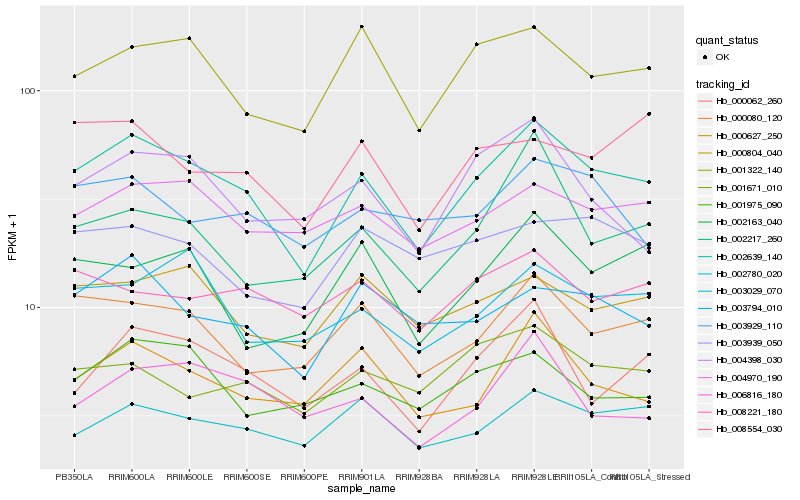
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002639_140 |
0.0 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 5 [Jatropha curcas] |
| 2 |
Hb_000080_120 |
0.0960234469 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 3 |
Hb_001322_140 |
0.1029908595 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002780_020 |
0.1057071084 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 [Jatropha curcas] |
| 5 |
Hb_003794_010 |
0.1069760227 |
- |
- |
PREDICTED: uncharacterized protein LOC105641013 [Jatropha curcas] |
| 6 |
Hb_000627_250 |
0.1073435409 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000062_260 |
0.1076617246 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 8 |
Hb_004970_190 |
0.1091994216 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_003029_070 |
0.1110513086 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000804_040 |
0.111325377 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_001975_090 |
0.1144125277 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 12 |
Hb_001671_010 |
0.1152626844 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 13 |
Hb_004398_030 |
0.1166725738 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 14 |
Hb_003929_110 |
0.1172336386 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_003939_050 |
0.1178779277 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_006816_180 |
0.1182614109 |
- |
- |
- |
| 17 |
Hb_002217_260 |
0.1186703093 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 18 |
Hb_002163_040 |
0.1196348062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_008554_030 |
0.1198644212 |
- |
- |
PREDICTED: dnaJ homolog 1, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_008221_180 |
0.1201301941 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |