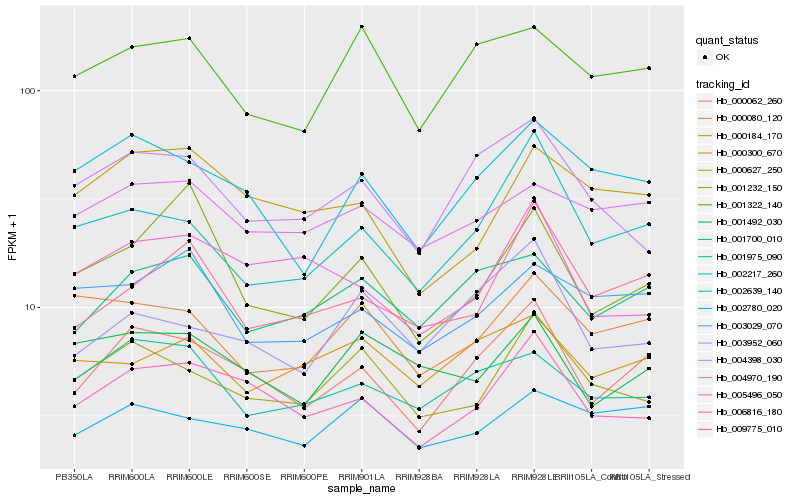
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 2 |
Hb_006816_180 |
0.0897314038 |
- |
- |
- |
| 3 |
Hb_002639_140 |
0.1076617246 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 5 [Jatropha curcas] |
| 4 |
Hb_001700_010 |
0.112746882 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 5 |
Hb_002217_260 |
0.1142053837 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 6 |
Hb_009775_010 |
0.1250456084 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 7 |
Hb_004398_030 |
0.1266241456 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 8 |
Hb_001975_090 |
0.1268522253 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 9 |
Hb_000627_250 |
0.1276115441 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003952_060 |
0.1278198872 |
- |
- |
hypothetical protein POPTR_0016s11000g [Populus trichocarpa] |
| 11 |
Hb_001322_140 |
0.1283670376 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000300_670 |
0.1317743828 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004970_190 |
0.1322387423 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_002780_020 |
0.1336909692 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 [Jatropha curcas] |
| 15 |
Hb_005496_050 |
0.1340497988 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 [Jatropha curcas] |
| 16 |
Hb_000080_120 |
0.1400140508 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350-like [Jatropha curcas] |
| 17 |
Hb_000184_170 |
0.1413090468 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 18 |
Hb_001492_030 |
0.1417728527 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18975, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001232_150 |
0.1419988008 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 20 |
Hb_003029_070 |
0.1424379824 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |