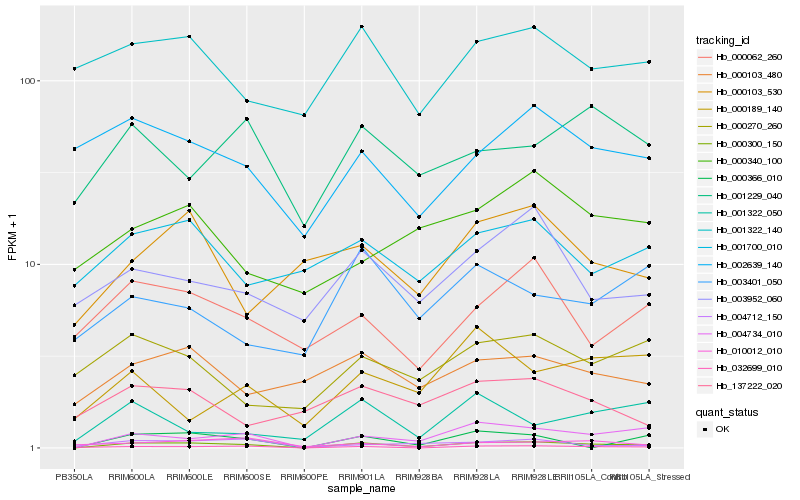
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_150 |
0.0 |
- |
- |
Peroxidase 44 precursor, putative [Ricinus communis] |
| 2 |
Hb_004734_010 |
0.1657818618 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 3 |
Hb_032699_010 |
0.2107879751 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_000366_010 |
0.2430322782 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g08070-like, partial [Pyrus x bretschneideri] |
| 5 |
Hb_004712_150 |
0.2516765417 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_000103_480 |
0.2628727232 |
- |
- |
- |
| 7 |
Hb_003952_060 |
0.2630135877 |
- |
- |
hypothetical protein POPTR_0016s11000g [Populus trichocarpa] |
| 8 |
Hb_000062_260 |
0.2638724681 |
- |
- |
PREDICTED: uncharacterized protein LOC105644756 [Jatropha curcas] |
| 9 |
Hb_001322_140 |
0.2646466429 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000270_260 |
0.2667409351 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |
| 11 |
Hb_002639_140 |
0.2684431725 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 5 [Jatropha curcas] |
| 12 |
Hb_001229_040 |
0.2691690564 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 13 |
Hb_010012_010 |
0.2702551227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001322_050 |
0.2704019128 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16835, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000103_530 |
0.2721448604 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 16 |
Hb_001700_010 |
0.2724771641 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 17 |
Hb_003401_050 |
0.2745152006 |
- |
- |
PREDICTED: origin of replication complex subunit 2 [Jatropha curcas] |
| 18 |
Hb_000340_100 |
0.2753267899 |
- |
- |
PREDICTED: protein PALE CRESS, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000189_140 |
0.2798004534 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_137222_020 |
0.2801982732 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |