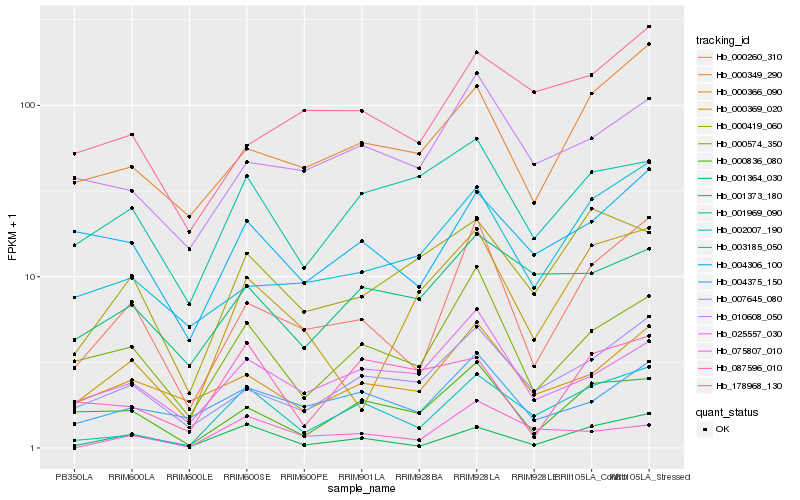
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002007_190 |
0.0 |
- |
- |
hypothetical protein RCOM_1432220 [Ricinus communis] |
| 2 |
Hb_007645_080 |
0.1947293783 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 3 |
Hb_001364_030 |
0.2005359755 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 4 |
Hb_000260_310 |
0.2028851906 |
- |
- |
PREDICTED: uncharacterized protein LOC105649060 [Jatropha curcas] |
| 5 |
Hb_001373_180 |
0.2066994012 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000349_290 |
0.2101546235 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |
| 7 |
Hb_000574_350 |
0.2106553126 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 8 |
Hb_000419_060 |
0.2107099411 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 9 |
Hb_004375_150 |
0.2204156316 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 10 |
Hb_010608_050 |
0.2204825024 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 11 |
Hb_000836_080 |
0.2213748208 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g46050, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001969_090 |
0.2214621398 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 13 |
Hb_087596_010 |
0.2226656144 |
- |
- |
PREDICTED: disease resistance protein RPS4-like [Jatropha curcas] |
| 14 |
Hb_025557_030 |
0.2233646192 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_075807_010 |
0.2234258722 |
- |
- |
PREDICTED: uncharacterized protein LOC105631775 [Jatropha curcas] |
| 16 |
Hb_004306_100 |
0.2234992046 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |
| 17 |
Hb_000369_020 |
0.2236951714 |
- |
- |
PREDICTED: probable fructokinase-7 [Jatropha curcas] |
| 18 |
Hb_003185_050 |
0.2244332389 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 19 |
Hb_178968_130 |
0.2248254267 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 20 |
Hb_000366_090 |
0.225710414 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |