| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
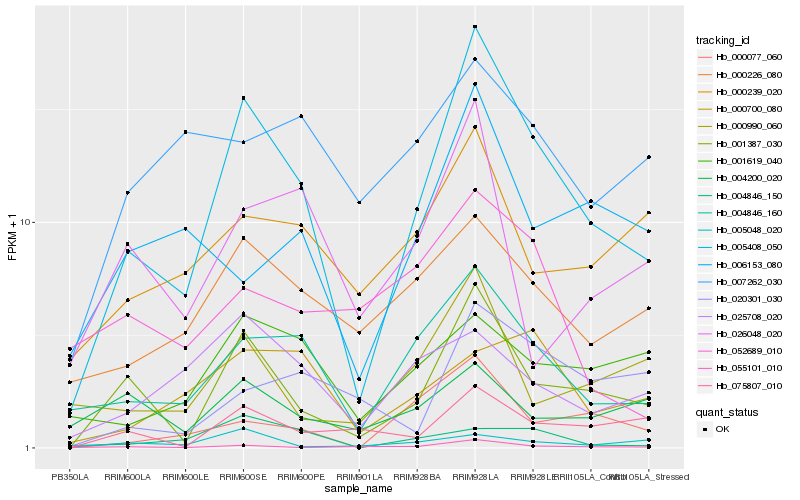
Hb_005408_050 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 2 |
Hb_001387_030 |
0.1642038431 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0013s13920g [Populus trichocarpa] |
| 3 |
Hb_004846_160 |
0.1885803024 |
- |
- |
- |
| 4 |
Hb_055101_010 |
0.2376736547 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000077_060 |
0.2460682536 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 6 |
Hb_000226_080 |
0.2509641262 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_075807_010 |
0.2626268932 |
- |
- |
PREDICTED: uncharacterized protein LOC105631775 [Jatropha curcas] |
| 8 |
Hb_000990_060 |
0.2642586143 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 9 |
Hb_000239_020 |
0.2695529948 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 10 |
Hb_001619_040 |
0.2734761027 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 11 |
Hb_025708_020 |
0.2752852562 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_026048_020 |
0.2786498693 |
- |
- |
PREDICTED: uncharacterized protein LOC105645738 [Jatropha curcas] |
| 13 |
Hb_006153_080 |
0.2788743395 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |
| 14 |
Hb_004846_150 |
0.2804483029 |
- |
- |
hypothetical protein JCGZ_26968 [Jatropha curcas] |
| 15 |
Hb_000700_080 |
0.2854614463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_020301_030 |
0.287886432 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_007262_030 |
0.296305842 |
- |
- |
- |
| 18 |
Hb_052689_010 |
0.2964258654 |
- |
- |
heat shock cognate protein [Solanum commersonii] |
| 19 |
Hb_005048_020 |
0.2966168947 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 20 |
Hb_004200_020 |
0.2998589661 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |