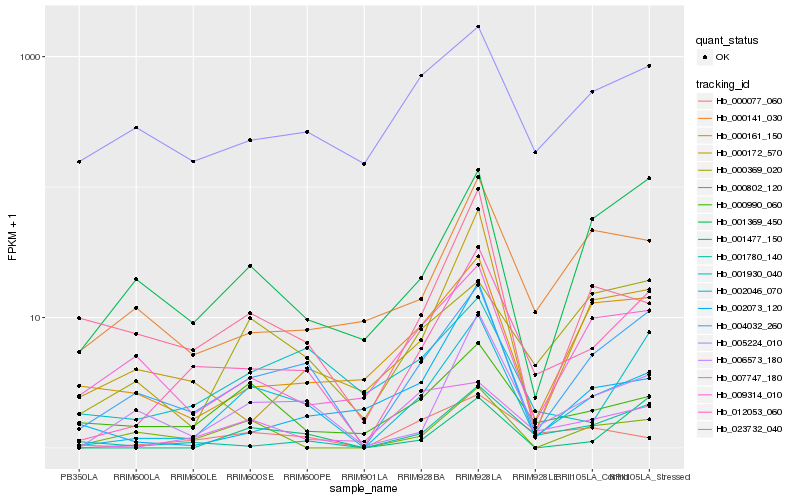
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002046_070 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like isoform X1 [Populus euphratica] |
| 2 |
Hb_004032_260 |
0.1703971706 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |
| 3 |
Hb_012053_060 |
0.1734146153 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g44820 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006573_180 |
0.1850917801 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1581940 [Ricinus communis] |
| 5 |
Hb_023732_040 |
0.2400935118 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_000990_060 |
0.2418361207 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 7 |
Hb_000077_060 |
0.2487469218 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 8 |
Hb_000161_150 |
0.2599030821 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 9 |
Hb_000172_570 |
0.2635149881 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009314_010 |
0.2656873746 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 11 |
Hb_000802_120 |
0.2671207115 |
- |
- |
- |
| 12 |
Hb_001369_450 |
0.2673921346 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_001930_040 |
0.2695795806 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_001477_150 |
0.2714760149 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000141_030 |
0.2723721147 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 16 |
Hb_005224_010 |
0.2736945713 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 17 |
Hb_002073_120 |
0.275489434 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein [Theobroma cacao] |
| 18 |
Hb_000369_020 |
0.2760223346 |
- |
- |
PREDICTED: probable fructokinase-7 [Jatropha curcas] |
| 19 |
Hb_001780_140 |
0.2761772237 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_007747_180 |
0.2780948965 |
transcription factor |
TF Family: TCP |
PREDICTED: bifunctional nitrilase/nitrile hydratase NIT4B-like [Jatropha curcas] |