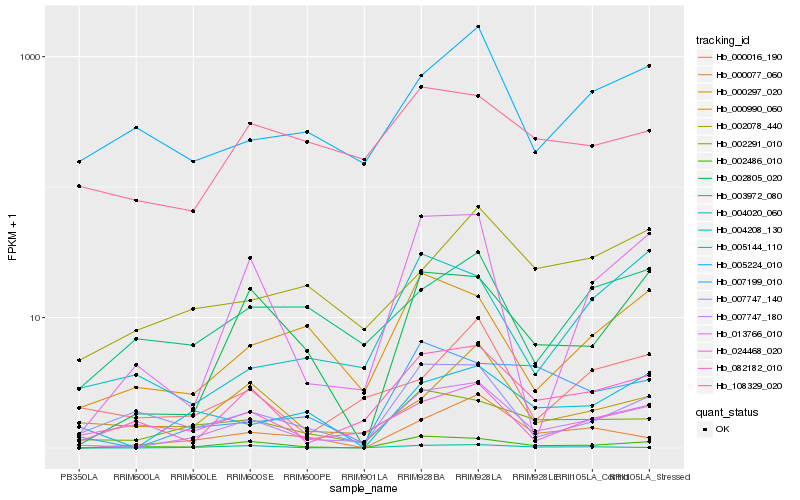
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007747_180 |
0.0 |
transcription factor |
TF Family: TCP |
PREDICTED: bifunctional nitrilase/nitrile hydratase NIT4B-like [Jatropha curcas] |
| 2 |
Hb_082182_010 |
0.1662636179 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 3 |
Hb_002486_010 |
0.1872951485 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 1 [Jatropha curcas] |
| 4 |
Hb_002291_010 |
0.1990745893 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 5 |
Hb_013766_010 |
0.2071827794 |
- |
- |
hypothetical protein JCGZ_00483 [Jatropha curcas] |
| 6 |
Hb_002805_020 |
0.2096946576 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000297_020 |
0.2219189503 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 8 |
Hb_005224_010 |
0.2256530941 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 9 |
Hb_024468_020 |
0.22658666 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 10 |
Hb_004020_060 |
0.2288179046 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_007747_140 |
0.2289362815 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_005144_110 |
0.2315715834 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 13 |
Hb_000077_060 |
0.2334350033 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH36-like [Jatropha curcas] |
| 14 |
Hb_000990_060 |
0.2398843566 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 15 |
Hb_004208_130 |
0.2420276776 |
- |
- |
hypothetical protein JCGZ_16532 [Jatropha curcas] |
| 16 |
Hb_007199_010 |
0.2431418901 |
- |
- |
PREDICTED: protein kinase APK1A, chloroplastic [Nelumbo nucifera] |
| 17 |
Hb_000016_190 |
0.2437856143 |
- |
- |
ABC transporter B family member 28 [Glycine soja] |
| 18 |
Hb_002078_440 |
0.2458344056 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003972_080 |
0.2458581188 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 20 |
Hb_108329_020 |
0.2473384639 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |