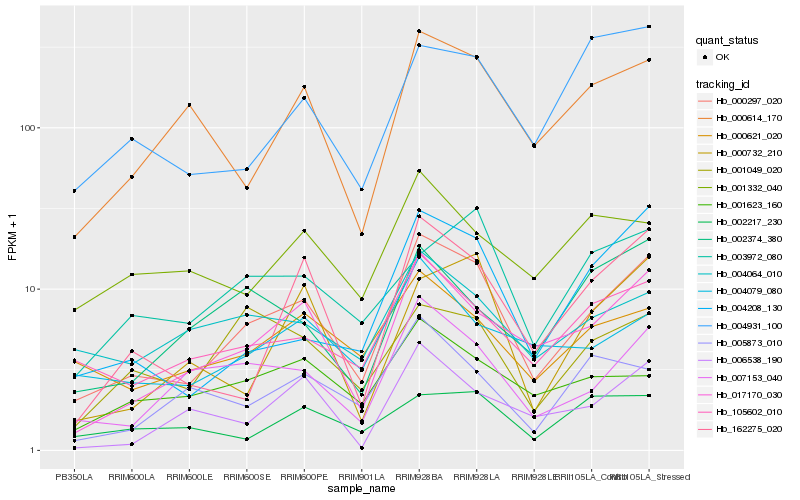
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000297_020 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 2 |
Hb_017170_030 |
0.1350327929 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 3 |
Hb_007153_040 |
0.1524143344 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 4 |
Hb_105602_010 |
0.1539927905 |
- |
- |
Ketose-bisphosphate aldolase class-II family protein isoform 5 [Theobroma cacao] |
| 5 |
Hb_004208_130 |
0.1632289568 |
- |
- |
hypothetical protein JCGZ_16532 [Jatropha curcas] |
| 6 |
Hb_162275_020 |
0.1652940385 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 7 |
Hb_000614_170 |
0.1729607175 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 8 |
Hb_000621_020 |
0.173622502 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 9 |
Hb_004079_080 |
0.1827209887 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 10 |
Hb_004064_010 |
0.1849733859 |
- |
- |
PREDICTED: uncharacterized protein LOC105133340 isoform X1 [Populus euphratica] |
| 11 |
Hb_001049_020 |
0.1875738115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001623_160 |
0.1920221268 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 13 |
Hb_000732_210 |
0.1924558134 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 14 |
Hb_004931_100 |
0.1933411271 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |
| 15 |
Hb_003972_080 |
0.1935108871 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 16 |
Hb_006538_190 |
0.1974474853 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002374_380 |
0.1975655219 |
- |
- |
Acyl-CoA N-acyltransferases (NAT) superfamily protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_002217_230 |
0.2020402417 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 19 |
Hb_005873_010 |
0.2022777588 |
- |
- |
cysteine desulfurylase, putative [Ricinus communis] |
| 20 |
Hb_001332_040 |
0.2023228168 |
- |
- |
PREDICTED: transcription initiation factor IIB [Fragaria vesca subsp. vesca] |