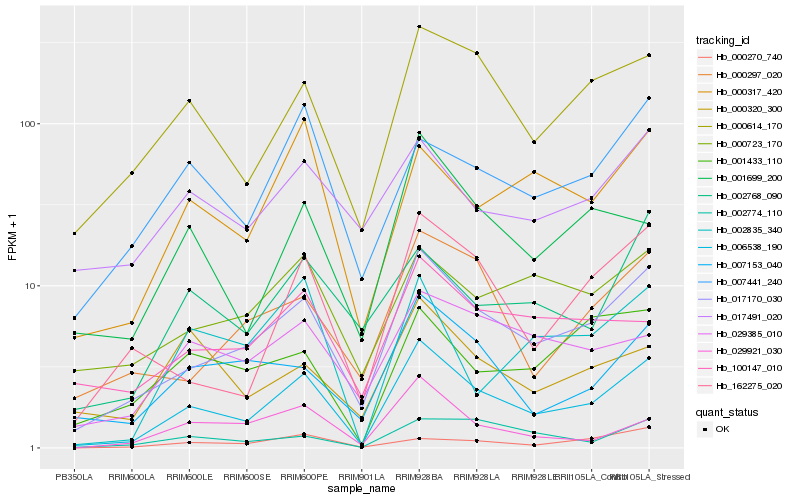
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006538_190 |
0.0 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_017170_030 |
0.1003496446 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 3 |
Hb_000614_170 |
0.1628925306 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 4 |
Hb_162275_020 |
0.1753155494 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 5 |
Hb_001699_200 |
0.1875473934 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 6 |
Hb_007153_040 |
0.1888210516 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000317_420 |
0.1895692438 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 8 |
Hb_002835_340 |
0.1972072088 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 9 |
Hb_000297_020 |
0.1974474853 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 10 |
Hb_029921_030 |
0.1987887413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007441_240 |
0.1994087421 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 12 |
Hb_002774_110 |
0.2007174607 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_029385_010 |
0.2013697101 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 14 |
Hb_002768_090 |
0.2039122954 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 15 |
Hb_000723_170 |
0.2053473539 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 16 |
Hb_100147_010 |
0.2065917235 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 17 |
Hb_000270_740 |
0.2081530017 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 18 |
Hb_001433_110 |
0.2085816102 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |
| 19 |
Hb_017491_020 |
0.2126449399 |
- |
- |
hypothetical protein B456_011G056700 [Gossypium raimondii] |
| 20 |
Hb_000320_300 |
0.2145779553 |
- |
- |
prephenate dehydratase, putative [Ricinus communis] |