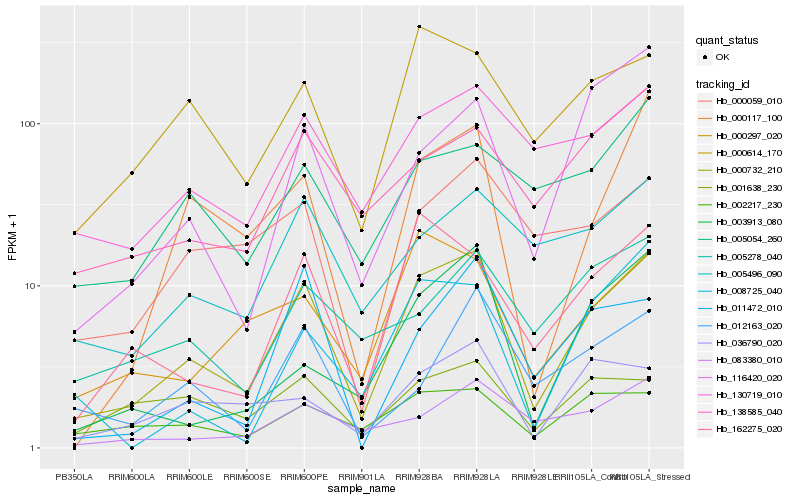
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_210 |
0.0 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 2 |
Hb_162275_020 |
0.1783497057 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 3 |
Hb_130719_010 |
0.189143159 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 4 |
Hb_000117_100 |
0.1899533846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000614_170 |
0.1900257465 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase-2C cytosolic [Gossypium arboreum] |
| 6 |
Hb_000297_020 |
0.1924558134 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 7 |
Hb_002217_230 |
0.1973968202 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 8 |
Hb_005496_090 |
0.1975934861 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_138585_040 |
0.2001354077 |
- |
- |
PREDICTED: proteasome subunit alpha type-4 [Jatropha curcas] |
| 10 |
Hb_083380_010 |
0.20100564 |
- |
- |
- |
| 11 |
Hb_036790_020 |
0.2021794043 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 12 |
Hb_000059_010 |
0.2038276288 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_011472_010 |
0.2058851373 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 14 |
Hb_003913_080 |
0.2088298704 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 15 |
Hb_008725_040 |
0.2091010778 |
- |
- |
PREDICTED: uncharacterized protein At4g29660 [Nelumbo nucifera] |
| 16 |
Hb_116420_020 |
0.2105413479 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 17 |
Hb_012163_020 |
0.2106282377 |
- |
- |
PREDICTED: uncharacterized protein LOC105650815 isoform X3 [Jatropha curcas] |
| 18 |
Hb_005054_260 |
0.2136085669 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A [Jatropha curcas] |
| 19 |
Hb_001638_230 |
0.2155593842 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 20 |
Hb_005278_040 |
0.2180235931 |
- |
- |
PREDICTED: sorcin-like isoform X1 [Populus euphratica] |