| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
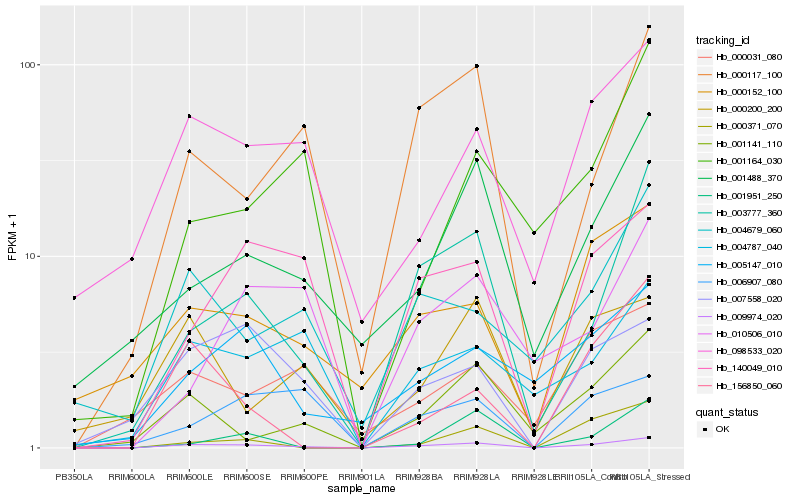
Hb_009974_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104427753 [Eucalyptus grandis] |
| 2 |
Hb_003777_360 |
0.1873396156 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_000117_100 |
0.2162316825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_140049_010 |
0.2216411678 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 5 |
Hb_001141_110 |
0.2245456983 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 6 |
Hb_007558_020 |
0.2278954154 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 7 |
Hb_001488_370 |
0.231375476 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 8 |
Hb_098533_020 |
0.2323180008 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 9 |
Hb_000152_100 |
0.2370209219 |
- |
- |
PREDICTED: probable protein phosphatase 2C 75 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004787_040 |
0.2412722227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004679_060 |
0.241674501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005147_010 |
0.2475705408 |
- |
- |
PREDICTED: uncharacterized protein LOC105629670 [Jatropha curcas] |
| 13 |
Hb_006907_080 |
0.2529822461 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 14 |
Hb_000371_070 |
0.2547810455 |
- |
- |
PREDICTED: uncharacterized protein LOC103863319 [Brassica rapa] |
| 15 |
Hb_000031_080 |
0.255446713 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 16 |
Hb_156850_060 |
0.2604749152 |
- |
- |
PREDICTED: uncharacterized protein LOC105635248 [Jatropha curcas] |
| 17 |
Hb_001951_250 |
0.2663169827 |
- |
- |
- |
| 18 |
Hb_000200_200 |
0.268056111 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 19 |
Hb_010506_010 |
0.2704182469 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 20 |
Hb_001164_030 |
0.2718591282 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |