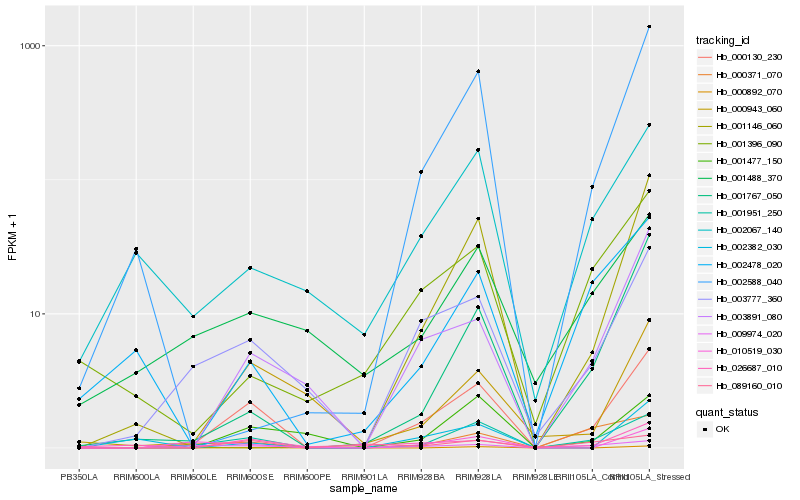
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000130_230 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 2 |
Hb_001951_250 |
0.1434588202 |
- |
- |
- |
| 3 |
Hb_003777_360 |
0.205348851 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_003891_080 |
0.2188624291 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |
| 5 |
Hb_001477_150 |
0.2512903555 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001767_050 |
0.2612287845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000892_070 |
0.2634439993 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_002067_140 |
0.2750360283 |
- |
- |
- |
| 9 |
Hb_000943_060 |
0.2780270522 |
- |
- |
PREDICTED: uncharacterized protein LOC105635589 [Jatropha curcas] |
| 10 |
Hb_010519_030 |
0.2822864652 |
- |
- |
- |
| 11 |
Hb_026687_010 |
0.2869575119 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 12 |
Hb_000371_070 |
0.2881885834 |
- |
- |
PREDICTED: uncharacterized protein LOC103863319 [Brassica rapa] |
| 13 |
Hb_002588_040 |
0.291207065 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 14 |
Hb_009974_020 |
0.2931651142 |
- |
- |
PREDICTED: uncharacterized protein LOC104427753 [Eucalyptus grandis] |
| 15 |
Hb_001146_060 |
0.2933229891 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 16 |
Hb_001488_370 |
0.2935059973 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 17 |
Hb_089160_010 |
0.2957121808 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 18 |
Hb_001396_090 |
0.2965620202 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 19 |
Hb_002382_030 |
0.2969019319 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Malus domestica] |
| 20 |
Hb_002478_020 |
0.2971460103 |
- |
- |
hypothetical protein POPTR_0010s19120g [Populus trichocarpa] |