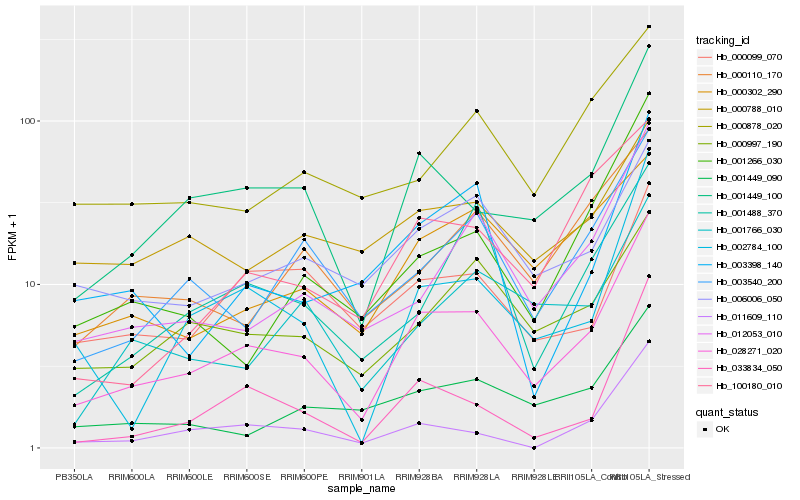
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028271_020 |
0.0 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 2 |
Hb_001449_100 |
0.1162625252 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 3 |
Hb_002784_100 |
0.1516571989 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 4 |
Hb_012053_010 |
0.157656389 |
- |
- |
srpk, putative [Ricinus communis] |
| 5 |
Hb_003540_200 |
0.1584855781 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 6 |
Hb_033834_050 |
0.1738085447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000997_190 |
0.174207521 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like [Jatropha curcas] |
| 8 |
Hb_006006_050 |
0.1743644466 |
- |
- |
PREDICTED: methylthioribose kinase-like [Gossypium raimondii] |
| 9 |
Hb_001449_090 |
0.1746157367 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_100180_010 |
0.1771396398 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 11 |
Hb_001266_030 |
0.1773428139 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_011609_110 |
0.1787989638 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 13 |
Hb_000110_170 |
0.1789638869 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_003398_140 |
0.1807741062 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000302_290 |
0.1816786426 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 16 |
Hb_000878_020 |
0.1821589786 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 17 |
Hb_000099_070 |
0.1834572968 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 18 |
Hb_001766_030 |
0.1841447781 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 19 |
Hb_001488_370 |
0.1861098327 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 20 |
Hb_000788_010 |
0.1897384824 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |