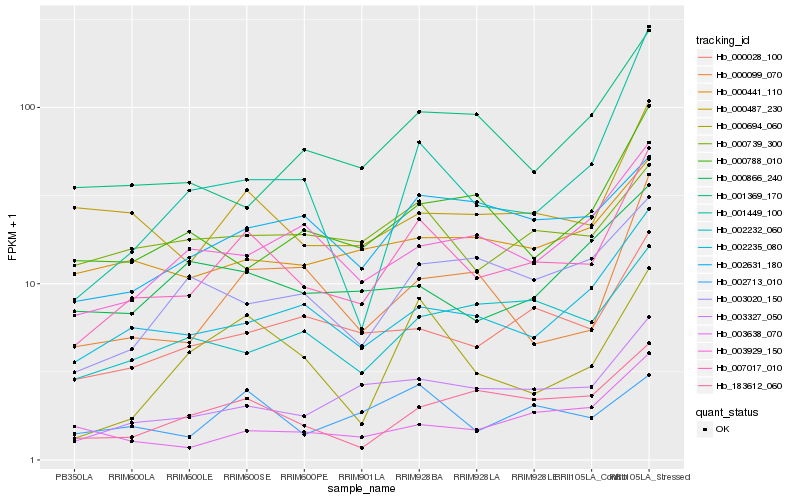
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007017_010 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 2 |
Hb_002235_080 |
0.1412839102 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 3 |
Hb_003327_050 |
0.1451163873 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 4 |
Hb_000028_100 |
0.1452189494 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 5 |
Hb_000487_230 |
0.1558815946 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 6 |
Hb_000099_070 |
0.1581967014 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 7 |
Hb_183612_060 |
0.1622063984 |
- |
- |
PREDICTED: uncharacterized protein LOC105632253 [Jatropha curcas] |
| 8 |
Hb_000694_060 |
0.1630126737 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 9 |
Hb_003929_150 |
0.1640231556 |
- |
- |
PREDICTED: rRNA-processing protein fcf2-like [Jatropha curcas] |
| 10 |
Hb_000441_110 |
0.1691326178 |
- |
- |
PREDICTED: uncharacterized protein LOC105646523 [Jatropha curcas] |
| 11 |
Hb_002631_180 |
0.1711863561 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 12 |
Hb_002713_010 |
0.1719307898 |
- |
- |
multidrug resistance protein 1, 2, putative [Ricinus communis] |
| 13 |
Hb_003020_150 |
0.1737499333 |
- |
- |
tropinone reductase, putative [Ricinus communis] |
| 14 |
Hb_000788_010 |
0.1745431614 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 15 |
Hb_002232_060 |
0.1762489044 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 16 |
Hb_001449_100 |
0.1779068873 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 17 |
Hb_000739_300 |
0.1779469891 |
- |
- |
PREDICTED: uncharacterized protein LOC105643092 [Jatropha curcas] |
| 18 |
Hb_001369_170 |
0.1816923769 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1-like [Gossypium raimondii] |
| 19 |
Hb_003638_070 |
0.1838151871 |
- |
- |
hypothetical protein EUGRSUZ_E00638 [Eucalyptus grandis] |
| 20 |
Hb_000866_240 |
0.1841986861 |
- |
- |
PREDICTED: 60S ribosomal protein L36-2-like [Jatropha curcas] |