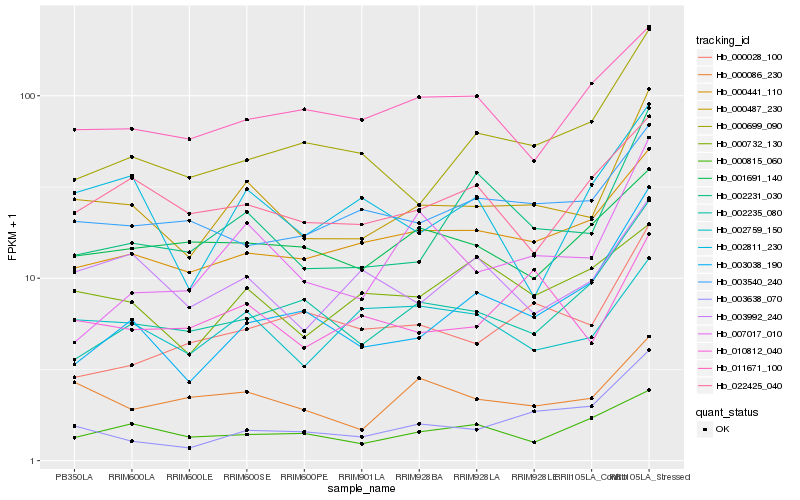
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000487_230 |
0.0 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 2 |
Hb_000441_110 |
0.1280569733 |
- |
- |
PREDICTED: uncharacterized protein LOC105646523 [Jatropha curcas] |
| 3 |
Hb_002231_030 |
0.1295576061 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 4 |
Hb_000086_230 |
0.1384586992 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000699_090 |
0.1425071396 |
- |
- |
cyclophilin, putative [Ricinus communis] |
| 6 |
Hb_010812_040 |
0.1463005408 |
transcription factor |
TF Family: SNF2 |
PREDICTED: E3 ubiquitin-protein ligase SHPRH [Jatropha curcas] |
| 7 |
Hb_003992_240 |
0.1487786211 |
- |
- |
- |
| 8 |
Hb_002235_080 |
0.1491319301 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 9 |
Hb_003638_070 |
0.1493636731 |
- |
- |
hypothetical protein EUGRSUZ_E00638 [Eucalyptus grandis] |
| 10 |
Hb_003540_240 |
0.1503063927 |
- |
- |
PREDICTED: ribosomal RNA-processing protein 17 [Jatropha curcas] |
| 11 |
Hb_002759_150 |
0.1519008165 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 22 isoform X1 [Jatropha curcas] |
| 12 |
Hb_022425_040 |
0.1526076463 |
- |
- |
PREDICTED: probable RNA methyltransferase At5g51130 [Jatropha curcas] |
| 13 |
Hb_007017_010 |
0.1558815946 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 14 |
Hb_002811_230 |
0.1582471186 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM23-1 [Jatropha curcas] |
| 15 |
Hb_000815_060 |
0.158282179 |
- |
- |
PREDICTED: uncharacterized protein LOC105630460 [Jatropha curcas] |
| 16 |
Hb_003038_190 |
0.1600926211 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105645961 [Jatropha curcas] |
| 17 |
Hb_000732_130 |
0.161324661 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_17108 [Jatropha curcas] |
| 18 |
Hb_000028_100 |
0.1613570774 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 19 |
Hb_011671_100 |
0.1619529932 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |
| 20 |
Hb_001691_140 |
0.162431591 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |