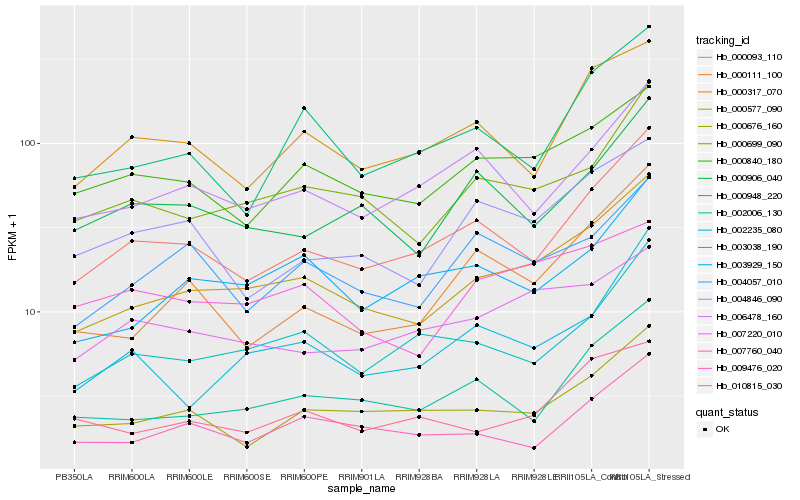
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000577_090 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000699_090 |
0.0951982533 |
- |
- |
cyclophilin, putative [Ricinus communis] |
| 3 |
Hb_000093_110 |
0.106258615 |
- |
- |
PREDICTED: cytochrome b5 isoform A [Jatropha curcas] |
| 4 |
Hb_002006_130 |
0.1118315247 |
- |
- |
PREDICTED: uncharacterized protein LOC105648712 [Jatropha curcas] |
| 5 |
Hb_000840_180 |
0.1131490377 |
- |
- |
protein BOLA2 [Jatropha curcas] |
| 6 |
Hb_003929_150 |
0.1156558048 |
- |
- |
PREDICTED: rRNA-processing protein fcf2-like [Jatropha curcas] |
| 7 |
Hb_000676_160 |
0.1204768307 |
- |
- |
folate carrier protein, putative [Ricinus communis] |
| 8 |
Hb_002235_080 |
0.1216736181 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 9 |
Hb_010815_030 |
0.1245884537 |
- |
- |
PREDICTED: (-)-germacrene D synthase [Vitis vinifera] |
| 10 |
Hb_004057_010 |
0.1259533416 |
- |
- |
PREDICTED: uncharacterized protein LOC105649313 [Jatropha curcas] |
| 11 |
Hb_000111_100 |
0.1265819866 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 12 |
Hb_000317_070 |
0.1283315935 |
- |
- |
PREDICTED: uncharacterized protein LOC103942205 isoform X2 [Pyrus x bretschneideri] |
| 13 |
Hb_009476_020 |
0.1283722523 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 14 |
Hb_006478_160 |
0.1284310632 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 15 |
Hb_003038_190 |
0.1290172207 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105645961 [Jatropha curcas] |
| 16 |
Hb_000948_220 |
0.1292474505 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |
| 17 |
Hb_004846_090 |
0.1298753962 |
transcription factor |
TF Family: HMG |
HMG-box DNA-binding family protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_007220_010 |
0.130332306 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase beta chain, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000906_040 |
0.1308047385 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007760_040 |
0.1309066184 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105630457 [Jatropha curcas] |