| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
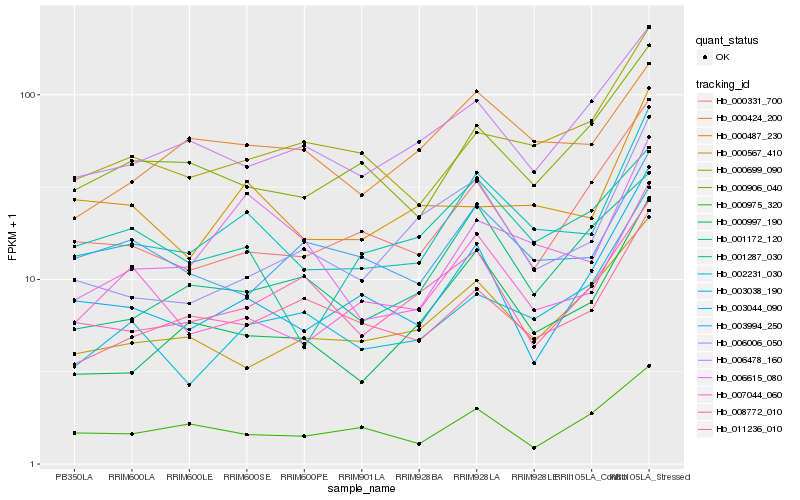
Hb_002231_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 2 |
Hb_007044_060 |
0.1133379159 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_003044_090 |
0.1265878264 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 4 |
Hb_000997_190 |
0.1277316007 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like [Jatropha curcas] |
| 5 |
Hb_000906_040 |
0.1281152028 |
- |
- |
PREDICTED: polynucleotide 5'-hydroxyl-kinase nol9 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000487_230 |
0.1295576061 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 7 |
Hb_000699_090 |
0.1303794139 |
- |
- |
cyclophilin, putative [Ricinus communis] |
| 8 |
Hb_006615_080 |
0.1304893627 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000331_700 |
0.1313159543 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 10 |
Hb_011236_010 |
0.1342747091 |
- |
- |
hypothetical protein JCGZ_17006 [Jatropha curcas] |
| 11 |
Hb_006478_160 |
0.1349294765 |
- |
- |
20S proteasome beta subunit D1 [Hevea brasiliensis] |
| 12 |
Hb_000567_410 |
0.1407210555 |
- |
- |
hypothetical protein JCGZ_12784 [Jatropha curcas] |
| 13 |
Hb_003994_250 |
0.1435600019 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 14 |
Hb_003038_190 |
0.1447490463 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105645961 [Jatropha curcas] |
| 15 |
Hb_000424_200 |
0.1457881466 |
- |
- |
hypothetical protein JCGZ_07583 [Jatropha curcas] |
| 16 |
Hb_006006_050 |
0.1470136467 |
- |
- |
PREDICTED: methylthioribose kinase-like [Gossypium raimondii] |
| 17 |
Hb_001287_030 |
0.1480447105 |
- |
- |
30S ribosomal protein S31, mitochondrial precursor, putative [Ricinus communis] |
| 18 |
Hb_000975_320 |
0.1484888009 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001172_120 |
0.1501239107 |
- |
- |
hypothetical protein B456_012G082000 [Gossypium raimondii] |
| 20 |
Hb_008772_010 |
0.1511504796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |