| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
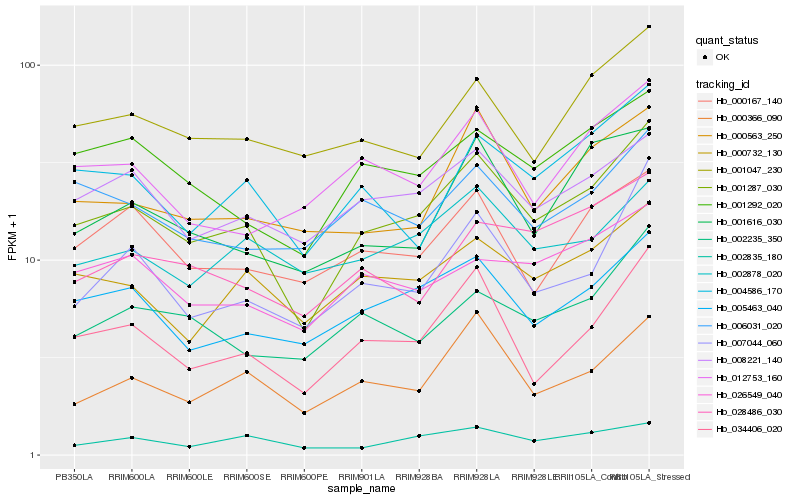
Hb_001287_030 |
0.0 |
- |
- |
30S ribosomal protein S31, mitochondrial precursor, putative [Ricinus communis] |
| 2 |
Hb_034406_020 |
0.0993366694 |
- |
- |
PREDICTED: jmjC domain-containing protein 7 [Jatropha curcas] |
| 3 |
Hb_005463_040 |
0.1028272779 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 4 |
Hb_000732_130 |
0.1083052218 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_17108 [Jatropha curcas] |
| 5 |
Hb_001292_020 |
0.1095081856 |
- |
- |
PREDICTED: keratin, type II cytoskeletal 5 isoform X1 [Sesamum indicum] |
| 6 |
Hb_008221_140 |
0.1106101308 |
- |
- |
PREDICTED: uncharacterized protein LOC105644226 [Jatropha curcas] |
| 7 |
Hb_004586_170 |
0.1121771692 |
- |
- |
hypothetical protein PRUPE_ppa011196mg [Prunus persica] |
| 8 |
Hb_000366_090 |
0.1131567525 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |
| 9 |
Hb_007044_060 |
0.1164268332 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_001047_230 |
0.1167812993 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_000563_250 |
0.1185186407 |
- |
- |
PREDICTED: uncharacterized protein LOC105643472 [Jatropha curcas] |
| 12 |
Hb_002235_350 |
0.1195304953 |
- |
- |
PREDICTED: uncharacterized protein LOC105635929 [Jatropha curcas] |
| 13 |
Hb_002835_180 |
0.119586243 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_002878_020 |
0.1217855197 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 15 |
Hb_000167_140 |
0.123019347 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 16 |
Hb_026549_040 |
0.1235497727 |
- |
- |
PREDICTED: protein SLOW GREEN 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_006031_020 |
0.1244982044 |
- |
- |
PREDICTED: protein FLX-like 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001616_030 |
0.1245450081 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor-like protein At4g13040 isoform X2 [Jatropha curcas] |
| 19 |
Hb_012753_160 |
0.1250393789 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 20 |
Hb_028486_030 |
0.125124583 |
- |
- |
hypothetical protein JCGZ_12706 [Jatropha curcas] |