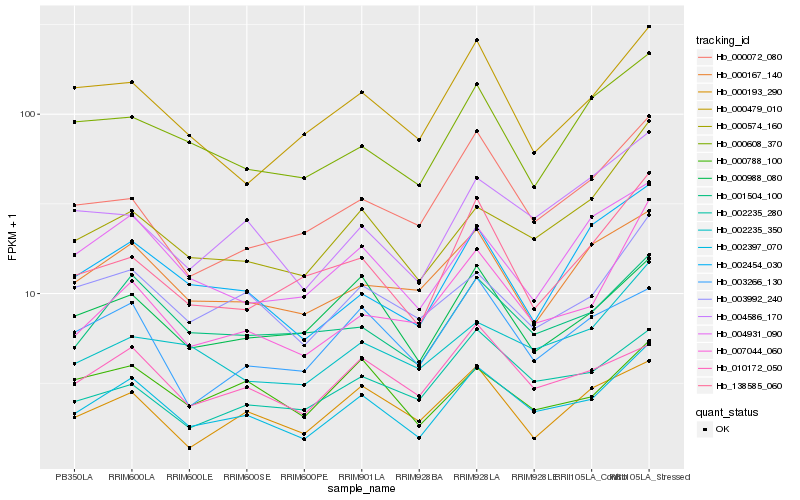
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002397_070 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 2 |
Hb_001504_100 |
0.0994704373 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_003992_240 |
0.1057804454 |
- |
- |
- |
| 4 |
Hb_007044_060 |
0.1098912399 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_004931_090 |
0.1123437718 |
- |
- |
PREDICTED: chromatin modification-related protein MEAF6-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_004586_170 |
0.1140544818 |
- |
- |
hypothetical protein PRUPE_ppa011196mg [Prunus persica] |
| 7 |
Hb_000574_160 |
0.1143493135 |
- |
- |
PREDICTED: arogenate dehydrogenase 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000988_080 |
0.115108669 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_010172_050 |
0.1168298779 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065 [Jatropha curcas] |
| 10 |
Hb_138585_060 |
0.117518997 |
- |
- |
PREDICTED: uncharacterized protein LOC105648743 [Jatropha curcas] |
| 11 |
Hb_000167_140 |
0.118581259 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 12 |
Hb_002454_030 |
0.1186603032 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B [Jatropha curcas] |
| 13 |
Hb_002235_280 |
0.1194380117 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15300 [Jatropha curcas] |
| 14 |
Hb_000072_080 |
0.1196378272 |
- |
- |
PREDICTED: probable ribosomal protein S11, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000608_370 |
0.1210722989 |
- |
- |
acyl-carrier-protein [Triadica sebifera] |
| 16 |
Hb_002235_350 |
0.1210808842 |
- |
- |
PREDICTED: uncharacterized protein LOC105635929 [Jatropha curcas] |
| 17 |
Hb_003266_130 |
0.1226432248 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 18 |
Hb_000479_010 |
0.1230172824 |
- |
- |
PREDICTED: 40S ribosomal protein S19-3-like [Eucalyptus grandis] |
| 19 |
Hb_000788_100 |
0.1243536139 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g62890-like [Jatropha curcas] |
| 20 |
Hb_000193_290 |
0.1244763728 |
- |
- |
hypothetical protein JCGZ_11661 [Jatropha curcas] |