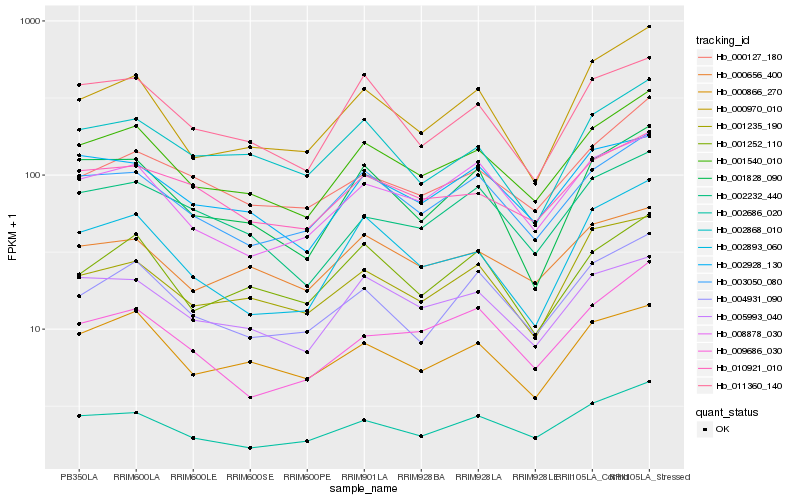
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001540_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639920 [Jatropha curcas] |
| 2 |
Hb_003050_080 |
0.0569994415 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 3 |
Hb_002686_020 |
0.0617098567 |
- |
- |
galactose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 4 |
Hb_008878_030 |
0.0731413053 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_004931_090 |
0.0739112449 |
- |
- |
PREDICTED: chromatin modification-related protein MEAF6-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_005993_040 |
0.0748387358 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit wdr4 [Jatropha curcas] |
| 7 |
Hb_000970_010 |
0.0751698281 |
- |
- |
PREDICTED: 60S ribosomal protein L26-1 [Jatropha curcas] |
| 8 |
Hb_002928_130 |
0.0752370183 |
- |
- |
PREDICTED: small nuclear ribonucleoprotein Sm D3 [Jatropha curcas] |
| 9 |
Hb_010921_010 |
0.0760985826 |
- |
- |
Small nuclear ribonucleoprotein family protein [Theobroma cacao] |
| 10 |
Hb_000127_180 |
0.0776584811 |
- |
- |
40S ribosomal protein S18, putative [Ricinus communis] |
| 11 |
Hb_002893_060 |
0.0783186448 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 50 [Jatropha curcas] |
| 12 |
Hb_000866_270 |
0.0784328365 |
- |
- |
PREDICTED: uncharacterized protein LOC105642993 [Jatropha curcas] |
| 13 |
Hb_011360_140 |
0.0801639341 |
- |
- |
40S ribosomal protein S19, putative [Ricinus communis] |
| 14 |
Hb_002232_440 |
0.0802754922 |
- |
- |
PREDICTED: uncharacterized protein LOC105130257 [Populus euphratica] |
| 15 |
Hb_001235_190 |
0.0802835521 |
- |
- |
PREDICTED: peroxisome biogenesis protein 19-2-like [Jatropha curcas] |
| 16 |
Hb_001828_090 |
0.082128188 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 17 |
Hb_000656_400 |
0.0827767329 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 18 |
Hb_001252_110 |
0.0830027736 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_009686_030 |
0.0850727711 |
- |
- |
- |
| 20 |
Hb_002868_010 |
0.0862440789 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |