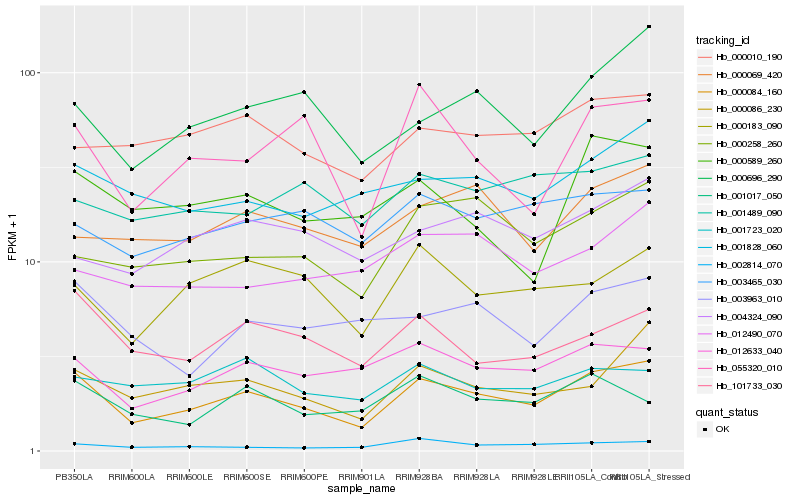
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000084_160 |
0.0 |
- |
- |
SGNH hydrolase-type esterase superfamily protein, putative [Theobroma cacao] |
| 2 |
Hb_000086_230 |
0.127051607 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 3 |
Hb_012633_040 |
0.1327576435 |
- |
- |
- |
| 4 |
Hb_101733_030 |
0.1363140616 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 5 |
Hb_001828_060 |
0.1373299567 |
desease resistance |
Gene Name: AAA_31 |
nucleotide-binding protein, putative [Ricinus communis] |
| 6 |
Hb_000696_290 |
0.1373323882 |
- |
- |
PREDICTED: UPF0664 stress-induced protein C29B12.11c [Jatropha curcas] |
| 7 |
Hb_002814_070 |
0.1398565578 |
- |
- |
PREDICTED: uncharacterized protein LOC104877459 [Vitis vinifera] |
| 8 |
Hb_055320_010 |
0.1402685057 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 9 |
Hb_000258_260 |
0.1405579185 |
- |
- |
PREDICTED: protein HGH1 homolog [Jatropha curcas] |
| 10 |
Hb_000183_090 |
0.1416367781 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003963_010 |
0.1424710075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000010_190 |
0.1467693347 |
- |
- |
PREDICTED: protein-L-isoaspartate O-methyltransferase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000069_420 |
0.1475440508 |
- |
- |
Beclin-1, putative [Ricinus communis] |
| 14 |
Hb_003465_030 |
0.1490671124 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000589_260 |
0.1497479913 |
- |
- |
3-hydroxybutyryl-CoA dehydratase, putative [Ricinus communis] |
| 16 |
Hb_001017_050 |
0.1523581322 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 17 |
Hb_004324_090 |
0.1536705885 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 18 |
Hb_012490_070 |
0.156004476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001489_090 |
0.1561630669 |
- |
- |
PREDICTED: uncharacterized protein LOC105649775 [Jatropha curcas] |
| 20 |
Hb_001723_020 |
0.1573778956 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |