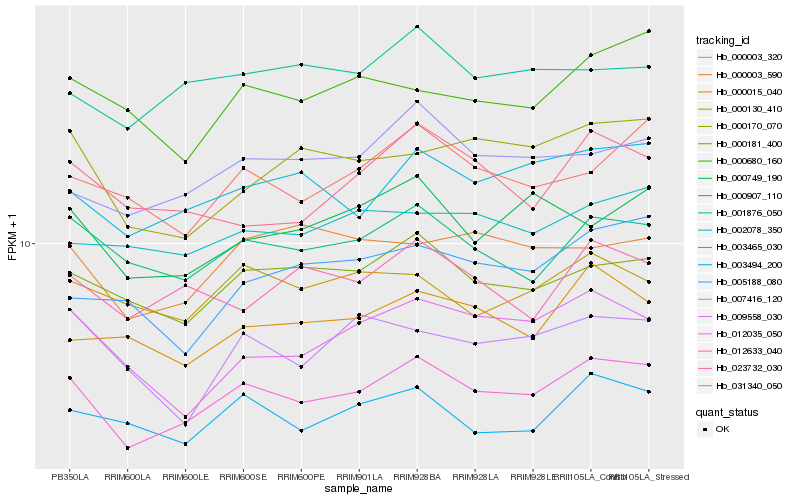
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012633_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_003465_030 |
0.0857274999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000907_110 |
0.0910634122 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 4 |
Hb_000181_400 |
0.0913031661 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 5 |
Hb_007416_120 |
0.0938142036 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 6 |
Hb_000170_070 |
0.0953320378 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |
| 7 |
Hb_000749_190 |
0.0967382887 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Populus euphratica] |
| 8 |
Hb_000003_320 |
0.0981743462 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 24 [Jatropha curcas] |
| 9 |
Hb_003494_200 |
0.0983925454 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 10 |
Hb_012035_050 |
0.0990968229 |
- |
- |
hypothetical protein JCGZ_10788 [Jatropha curcas] |
| 11 |
Hb_000015_040 |
0.1036667756 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X2 [Jatropha curcas] |
| 12 |
Hb_031340_050 |
0.103880226 |
- |
- |
PREDICTED: tropinone reductase 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001876_050 |
0.1042235861 |
- |
- |
hypothetical protein JCGZ_01286 [Jatropha curcas] |
| 14 |
Hb_023732_030 |
0.1048297642 |
- |
- |
PREDICTED: proteinaceous RNase P 2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_000003_590 |
0.1057540231 |
- |
- |
PREDICTED: uncharacterized protein LOC105631550 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000680_160 |
0.1059339162 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 17 |
Hb_002078_350 |
0.1071904801 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 18 |
Hb_005188_080 |
0.1074311715 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 19 |
Hb_000130_410 |
0.1075757042 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 20 |
Hb_009558_030 |
0.1081634038 |
- |
- |
myb3r3, putative [Ricinus communis] |