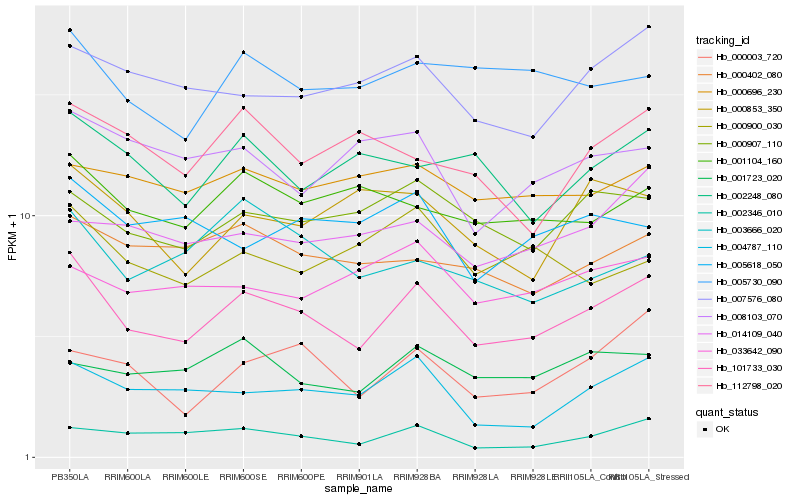
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101733_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 2 |
Hb_007576_080 |
0.1143984198 |
- |
- |
PREDICTED: uncharacterized protein LOC105643972 [Jatropha curcas] |
| 3 |
Hb_000907_110 |
0.1166436077 |
- |
- |
dihydroxyacetone kinase family protein [Populus trichocarpa] |
| 4 |
Hb_002346_010 |
0.1183404107 |
- |
- |
- |
| 5 |
Hb_004787_110 |
0.1184109362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000402_080 |
0.1191224585 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 7 |
Hb_001104_160 |
0.1193572838 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 8 |
Hb_005730_090 |
0.1215200368 |
- |
- |
PREDICTED: uncharacterized protein LOC105631484 [Jatropha curcas] |
| 9 |
Hb_001723_020 |
0.1254654659 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 10 |
Hb_002248_080 |
0.1267212465 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000003_720 |
0.1280718533 |
- |
- |
hypothetical protein POPTR_0003s04720g [Populus trichocarpa] |
| 12 |
Hb_003666_020 |
0.1298776131 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_000853_350 |
0.13000281 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 14 |
Hb_000696_230 |
0.1301689311 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 15 |
Hb_014109_040 |
0.1312501616 |
- |
- |
PREDICTED: probable folylpolyglutamate synthase [Jatropha curcas] |
| 16 |
Hb_005618_050 |
0.1320218829 |
- |
- |
PREDICTED: charged multivesicular body protein 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_033642_090 |
0.1322223948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_112798_020 |
0.1322877523 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 19 |
Hb_008103_070 |
0.1332477944 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 20 |
Hb_000900_030 |
0.1338761321 |
- |
- |
PREDICTED: WD repeat-containing protein 3 [Jatropha curcas] |