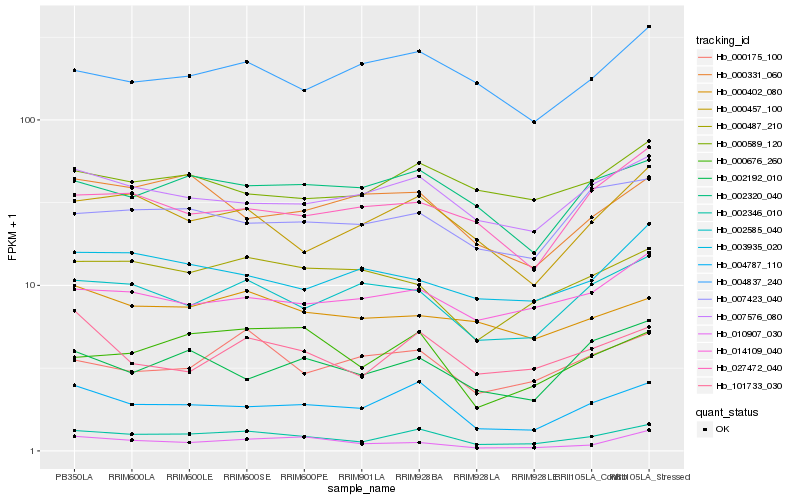
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002346_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004787_110 |
0.0829021197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000457_100 |
0.1045431888 |
- |
- |
prohibitin, putative [Ricinus communis] |
| 4 |
Hb_007576_080 |
0.1097638383 |
- |
- |
PREDICTED: uncharacterized protein LOC105643972 [Jatropha curcas] |
| 5 |
Hb_000676_260 |
0.112244522 |
- |
- |
PREDICTED: uncharacterized protein LOC105639070 [Jatropha curcas] |
| 6 |
Hb_002585_040 |
0.1143177766 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 7 |
Hb_002320_040 |
0.1168582324 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 8 |
Hb_101733_030 |
0.1183404107 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 9 |
Hb_000487_210 |
0.1228117503 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 10 |
Hb_007423_040 |
0.1237340717 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_014109_040 |
0.123789577 |
- |
- |
PREDICTED: probable folylpolyglutamate synthase [Jatropha curcas] |
| 12 |
Hb_000175_100 |
0.1249828326 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 13 |
Hb_000589_120 |
0.1260410976 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL30 [Gossypium raimondii] |
| 14 |
Hb_004837_240 |
0.1260978959 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 15 |
Hb_027472_040 |
0.1284872819 |
- |
- |
SKP1 [Hevea brasiliensis] |
| 16 |
Hb_010907_030 |
0.1293991587 |
- |
- |
- |
| 17 |
Hb_002192_010 |
0.1297243872 |
- |
- |
- |
| 18 |
Hb_003935_020 |
0.1299474988 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_000331_060 |
0.1306762562 |
- |
- |
PREDICTED: 60S ribosomal protein L17-2-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000402_080 |
0.1321588496 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |