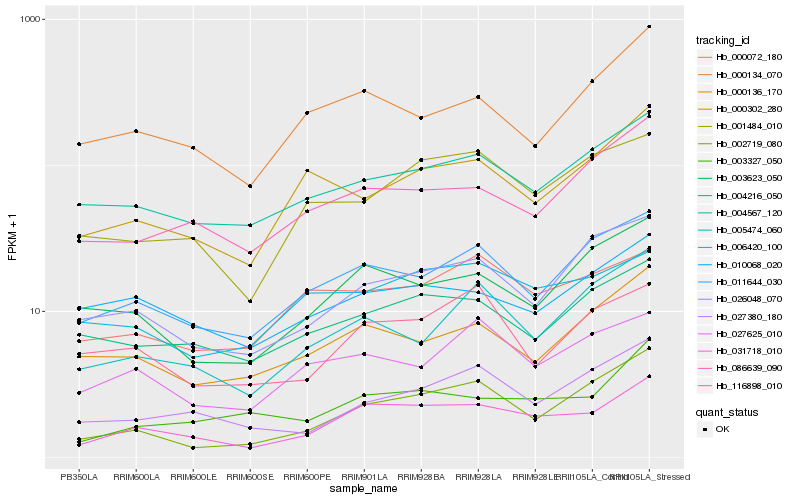
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031718_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_011644_030 |
0.129776672 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Pyrus x bretschneideri] |
| 3 |
Hb_004567_120 |
0.1389482051 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 4 |
Hb_003327_050 |
0.1398094781 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 5 |
Hb_003623_050 |
0.14125133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001484_010 |
0.1422627435 |
- |
- |
hypothetical protein POPTR_0005s07440g [Populus trichocarpa] |
| 7 |
Hb_026048_070 |
0.1438488565 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 8 |
Hb_002719_080 |
0.1456648761 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 9 |
Hb_086639_090 |
0.1470236358 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 10 |
Hb_004216_050 |
0.1477303719 |
- |
- |
PREDICTED: uncharacterized protein LOC105628422 [Jatropha curcas] |
| 11 |
Hb_027380_180 |
0.1505576128 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 12 |
Hb_000136_170 |
0.1524080279 |
- |
- |
PREDICTED: uncharacterized protein LOC105645501 [Jatropha curcas] |
| 13 |
Hb_000072_180 |
0.1525387254 |
- |
- |
PREDICTED: EVI5-like protein [Jatropha curcas] |
| 14 |
Hb_027625_010 |
0.1530886976 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 15 |
Hb_006420_100 |
0.1532360666 |
- |
- |
PREDICTED: protein bicaudal C homolog 1 [Jatropha curcas] |
| 16 |
Hb_000134_070 |
0.1541078554 |
- |
- |
60S ribosomal protein L6, putative [Ricinus communis] |
| 17 |
Hb_010068_020 |
0.1543475195 |
- |
- |
PREDICTED: uncharacterized protein LOC105632966 [Jatropha curcas] |
| 18 |
Hb_000302_280 |
0.1560504829 |
- |
- |
hypothetical protein ZEAMMB73_772347 [Zea mays] |
| 19 |
Hb_116898_010 |
0.1571243332 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005474_060 |
0.1572566342 |
- |
- |
hypothetical protein PHAVU_011G019700g [Phaseolus vulgaris] |