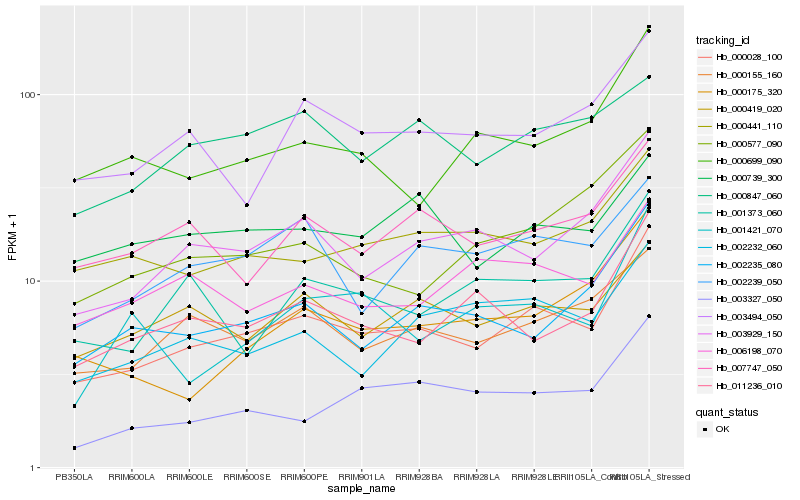
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_100 |
0.0 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 2 |
Hb_003929_150 |
0.1147268254 |
- |
- |
PREDICTED: rRNA-processing protein fcf2-like [Jatropha curcas] |
| 3 |
Hb_000155_160 |
0.1151117051 |
- |
- |
hypothetical protein POPTR_0011s05880g [Populus trichocarpa] |
| 4 |
Hb_002235_080 |
0.1220581262 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 5 |
Hb_003494_050 |
0.1238373441 |
- |
- |
hypothetical protein PRUPE_ppa013459mg [Prunus persica] |
| 6 |
Hb_000419_020 |
0.1242719856 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000441_110 |
0.1249822412 |
- |
- |
PREDICTED: uncharacterized protein LOC105646523 [Jatropha curcas] |
| 8 |
Hb_007747_050 |
0.1268014333 |
- |
- |
Peptidyl-prolyl cis-trans isomerase cypE, putative [Ricinus communis] |
| 9 |
Hb_002239_050 |
0.1277382306 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000699_090 |
0.1284067617 |
- |
- |
cyclophilin, putative [Ricinus communis] |
| 11 |
Hb_001373_060 |
0.1291615003 |
- |
- |
PREDICTED: structure-specific endonuclease subunit slx1 [Jatropha curcas] |
| 12 |
Hb_003327_050 |
0.1293731779 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 13 |
Hb_011236_010 |
0.1296151473 |
- |
- |
hypothetical protein JCGZ_17006 [Jatropha curcas] |
| 14 |
Hb_002232_060 |
0.1297204686 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 15 |
Hb_006198_070 |
0.134787492 |
- |
- |
pantoate-beta-alanine ligase, putative [Ricinus communis] |
| 16 |
Hb_000175_320 |
0.1358932202 |
- |
- |
PREDICTED: formin-like protein 14 [Jatropha curcas] |
| 17 |
Hb_000739_300 |
0.1397585823 |
- |
- |
PREDICTED: uncharacterized protein LOC105643092 [Jatropha curcas] |
| 18 |
Hb_000577_090 |
0.1406845801 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_001421_070 |
0.1424472546 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 20 |
Hb_000847_060 |
0.1428531933 |
- |
- |
hypothetical protein [Jatropha curcas] |