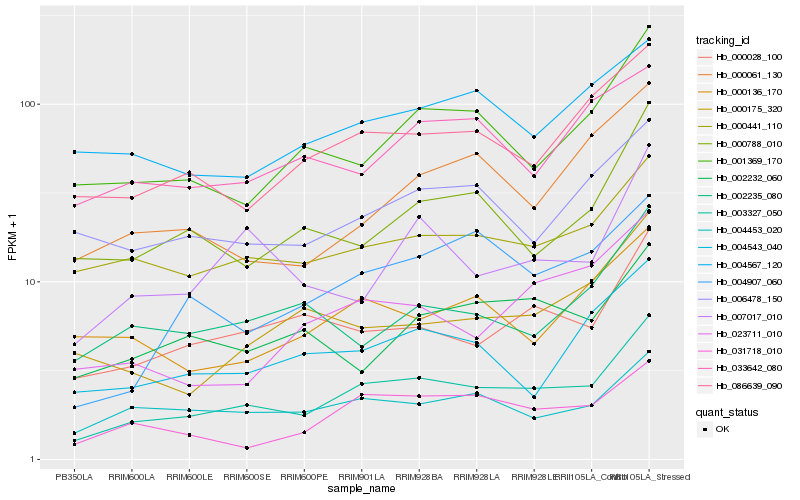
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003327_050 |
0.0 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 2 |
Hb_086639_090 |
0.127100851 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 3 |
Hb_000441_110 |
0.1279405235 |
- |
- |
PREDICTED: uncharacterized protein LOC105646523 [Jatropha curcas] |
| 4 |
Hb_000028_100 |
0.1293731779 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 5 |
Hb_001369_170 |
0.1378362674 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1-like [Gossypium raimondii] |
| 6 |
Hb_031718_010 |
0.1398094781 |
- |
- |
- |
| 7 |
Hb_004453_020 |
0.1420442278 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_004907_060 |
0.1420667147 |
- |
- |
PREDICTED: tropinone reductase homolog At5g06060-like [Jatropha curcas] |
| 9 |
Hb_004543_040 |
0.1427598077 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_006478_150 |
0.1430033433 |
- |
- |
mitochondrial ribosomal protein L24, putative [Ricinus communis] |
| 11 |
Hb_007017_010 |
0.1451163873 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 12 |
Hb_000788_010 |
0.1473268063 |
- |
- |
40S ribosomal protein S16A [Hevea brasiliensis] |
| 13 |
Hb_004567_120 |
0.1487272547 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 14 |
Hb_000175_320 |
0.1513029892 |
- |
- |
PREDICTED: formin-like protein 14 [Jatropha curcas] |
| 15 |
Hb_002232_060 |
0.1527749489 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 16 |
Hb_000136_170 |
0.1540886504 |
- |
- |
PREDICTED: uncharacterized protein LOC105645501 [Jatropha curcas] |
| 17 |
Hb_002235_080 |
0.1541367392 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 18 |
Hb_023711_010 |
0.154938638 |
- |
- |
- |
| 19 |
Hb_000061_130 |
0.1556413186 |
- |
- |
rotamase, putative [Ricinus communis] |
| 20 |
Hb_033642_080 |
0.1556582212 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |