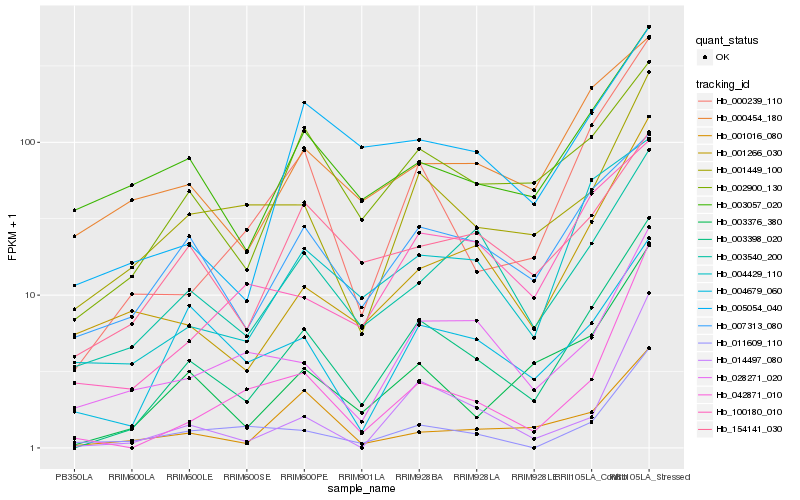
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003398_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 2 |
Hb_000239_110 |
0.147495128 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_003376_380 |
0.1563146357 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 4 |
Hb_004429_110 |
0.1744651374 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 5 |
Hb_007313_080 |
0.1749788128 |
- |
- |
hypothetical protein CICLE_v10002804mg [Citrus clementina] |
| 6 |
Hb_002900_130 |
0.1761251581 |
- |
- |
60S ribosomal L35a-3 -like protein [Gossypium arboreum] |
| 7 |
Hb_001449_100 |
0.1771203041 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 8 |
Hb_003540_200 |
0.1834449097 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 9 |
Hb_001266_030 |
0.1851572538 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_003057_020 |
0.1916187625 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 11 |
Hb_100180_010 |
0.1937314064 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 12 |
Hb_005054_040 |
0.1942623462 |
- |
- |
PREDICTED: 60S ribosomal protein L35a-1 [Gossypium raimondii] |
| 13 |
Hb_154141_030 |
0.195776882 |
- |
- |
PREDICTED: histone H2AX [Populus euphratica] |
| 14 |
Hb_000454_180 |
0.1976036026 |
- |
- |
PREDICTED: uncharacterized protein LOC105134496 [Populus euphratica] |
| 15 |
Hb_011609_110 |
0.1992862051 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 16 |
Hb_014497_080 |
0.1996486737 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 17 |
Hb_028271_020 |
0.199876983 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 18 |
Hb_001016_080 |
0.2016416723 |
- |
- |
hypothetical protein EUGRSUZ_J02452 [Eucalyptus grandis] |
| 19 |
Hb_042871_010 |
0.2016480613 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 20 |
Hb_004679_060 |
0.2033226657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |