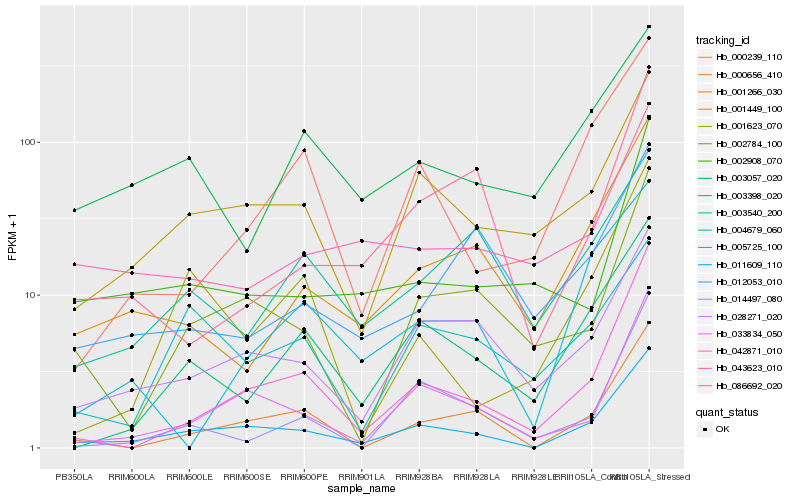
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011609_110 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 2 |
Hb_033834_050 |
0.1525034966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000656_410 |
0.1676065475 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 4 |
Hb_001449_100 |
0.1681413617 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 5 |
Hb_042871_010 |
0.1707153373 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 6 |
Hb_028271_020 |
0.1787989638 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 7 |
Hb_002784_100 |
0.1938914005 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 8 |
Hb_003398_020 |
0.1992862051 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 9 |
Hb_001266_030 |
0.2057268288 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000239_110 |
0.2185003611 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 11 |
Hb_001623_070 |
0.2191865339 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 12 |
Hb_002908_070 |
0.220971304 |
- |
- |
DNA gyrase subunit A, chloroplast/mitochondrial precursor, putative [Ricinus communis] |
| 13 |
Hb_014497_080 |
0.2300225564 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 14 |
Hb_005725_100 |
0.2305337287 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 15 |
Hb_043623_010 |
0.2310744021 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 16 |
Hb_003057_020 |
0.2326604305 |
- |
- |
40S ribosomal protein S23, putative [Ricinus communis] |
| 17 |
Hb_086692_020 |
0.2346844514 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_003540_200 |
0.2361665555 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 19 |
Hb_004679_060 |
0.2367286587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012053_010 |
0.2423191417 |
- |
- |
srpk, putative [Ricinus communis] |