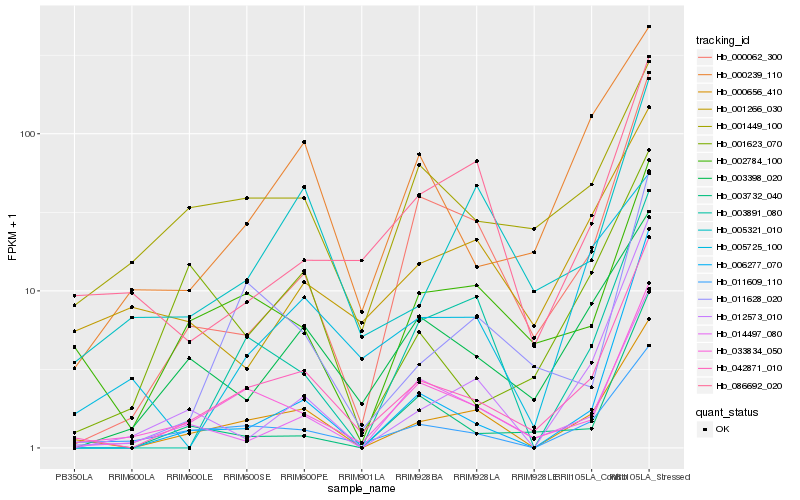
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042871_010 |
0.0 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 2 |
Hb_000656_410 |
0.1466217213 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 3 |
Hb_033834_050 |
0.1553090984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_011609_110 |
0.1707153373 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 5 |
Hb_000062_300 |
0.1764188461 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 6 |
Hb_000239_110 |
0.1880707806 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_014497_080 |
0.1928055895 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 8 |
Hb_006277_070 |
0.1935896698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011628_020 |
0.1968181444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002784_100 |
0.201014002 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 11 |
Hb_003732_040 |
0.2016418808 |
- |
- |
putative esterase family protein [Populus trichocarpa] |
| 12 |
Hb_003398_020 |
0.2016480613 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 13 |
Hb_012573_010 |
0.2127939605 |
- |
- |
PREDICTED: uncharacterized protein LOC105639338 [Jatropha curcas] |
| 14 |
Hb_005321_010 |
0.2177609456 |
- |
- |
PREDICTED: uncharacterized protein LOC105644542 [Jatropha curcas] |
| 15 |
Hb_003891_080 |
0.2215785106 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |
| 16 |
Hb_001623_070 |
0.2224313067 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 17 |
Hb_001266_030 |
0.2253860564 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_001449_100 |
0.2255377218 |
- |
- |
PREDICTED: transcription factor bHLH149 [Jatropha curcas] |
| 19 |
Hb_005725_100 |
0.2320874 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 20 |
Hb_086692_020 |
0.234712981 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |