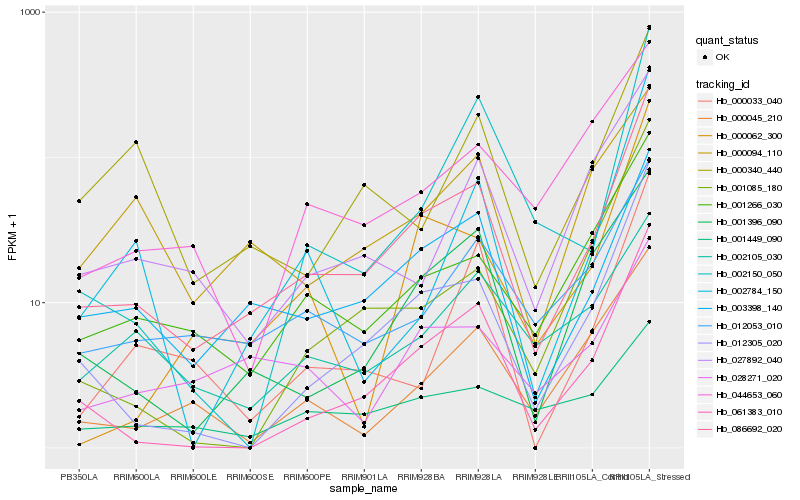
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086692_020 |
0.0 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_003398_140 |
0.1318869626 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_012305_020 |
0.1369396095 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 4 |
Hb_061383_010 |
0.144493189 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 5 |
Hb_001266_030 |
0.1526236807 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_012053_010 |
0.1631535949 |
- |
- |
srpk, putative [Ricinus communis] |
| 7 |
Hb_001396_090 |
0.1735120911 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 8 |
Hb_000045_210 |
0.1858262973 |
- |
- |
- |
| 9 |
Hb_000062_300 |
0.1874555649 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 10 |
Hb_027892_040 |
0.1887820618 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Jatropha curcas] |
| 11 |
Hb_028271_020 |
0.1913031764 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 12 |
Hb_044653_060 |
0.1999771442 |
- |
- |
PREDICTED: uncharacterized protein LOC105628750 [Jatropha curcas] |
| 13 |
Hb_000033_040 |
0.2011169578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002150_050 |
0.2029193587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000340_440 |
0.2029814026 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 16 |
Hb_002105_030 |
0.2037541433 |
- |
- |
- |
| 17 |
Hb_001085_180 |
0.2039170209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001449_090 |
0.204573613 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002784_150 |
0.2062497808 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 20 |
Hb_000094_110 |
0.2069199798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |