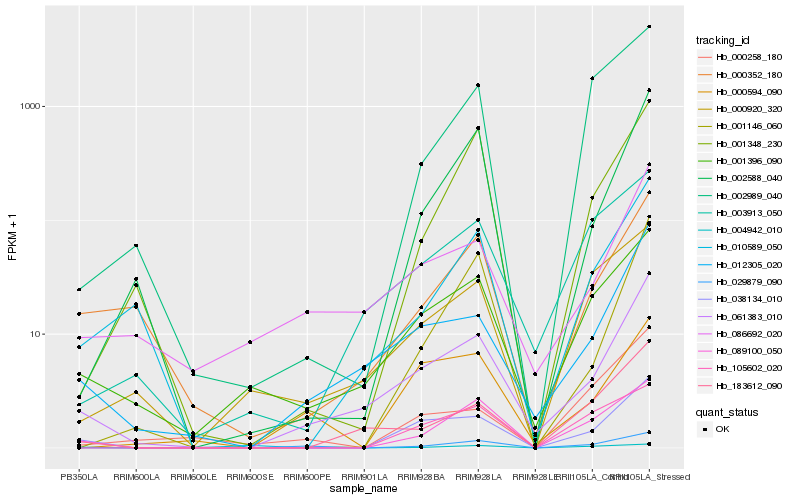
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038134_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_061383_010 |
0.1746553087 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 3 |
Hb_089100_050 |
0.1910952338 |
- |
- |
- |
| 4 |
Hb_001396_090 |
0.1982481847 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Jatropha curcas] |
| 5 |
Hb_012305_020 |
0.205895244 |
- |
- |
beta-1,3-glucanase [Hevea brasiliensis] |
| 6 |
Hb_105602_020 |
0.2151853186 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 7 |
Hb_000594_090 |
0.2258430307 |
- |
- |
PREDICTED: allene oxide cyclase 3, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_002588_040 |
0.2281468325 |
- |
- |
Pathogenesis-related protein 1 [Theobroma cacao] |
| 9 |
Hb_004942_010 |
0.2323622851 |
- |
- |
hypothetical protein AMTR_s00057p00201180 [Amborella trichopoda] |
| 10 |
Hb_000352_180 |
0.2330701104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010589_050 |
0.2340470467 |
- |
- |
- |
| 12 |
Hb_183612_090 |
0.2352531385 |
- |
- |
- |
| 13 |
Hb_002989_040 |
0.2448504298 |
- |
- |
hypothetical protein 1 [Hevea brasiliensis] |
| 14 |
Hb_003913_050 |
0.249056391 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 15 |
Hb_086692_020 |
0.2512446567 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_001146_060 |
0.2516709942 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 17 |
Hb_001348_230 |
0.2546848937 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000920_320 |
0.2550462698 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 19 |
Hb_000258_180 |
0.2580310743 |
- |
- |
PREDICTED: glutathione S-transferase U10-like [Vitis vinifera] |
| 20 |
Hb_029879_090 |
0.2592258817 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070 [Jatropha curcas] |