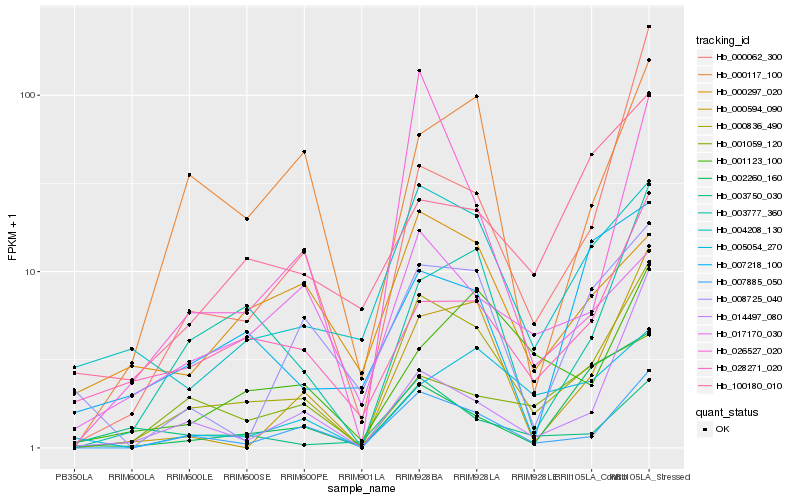
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000836_490 |
0.0 |
- |
- |
PREDICTED: E3 SUMO-protein ligase pli1-like [Jatropha curcas] |
| 2 |
Hb_007885_050 |
0.1429611715 |
- |
- |
PREDICTED: lignin-forming anionic peroxidase-like [Jatropha curcas] |
| 3 |
Hb_003777_360 |
0.1967879422 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_000594_090 |
0.2031361721 |
- |
- |
PREDICTED: allene oxide cyclase 3, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_004208_130 |
0.2046815551 |
- |
- |
hypothetical protein JCGZ_16532 [Jatropha curcas] |
| 6 |
Hb_028271_020 |
0.2218714988 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 7 |
Hb_000117_100 |
0.2301732243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_026527_020 |
0.2399937041 |
- |
- |
PREDICTED: putative methyltransferase DDB_G0268948 [Jatropha curcas] |
| 9 |
Hb_002260_160 |
0.2405396315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008725_040 |
0.2420679378 |
- |
- |
PREDICTED: uncharacterized protein At4g29660 [Nelumbo nucifera] |
| 11 |
Hb_005054_270 |
0.244201827 |
- |
- |
- |
| 12 |
Hb_001123_100 |
0.2468223386 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001059_120 |
0.2472057686 |
- |
- |
NADH dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_100180_010 |
0.2506933126 |
- |
- |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 15 |
Hb_003750_030 |
0.2530604666 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 16 |
Hb_014497_080 |
0.2558529109 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 17 |
Hb_000062_300 |
0.2561500999 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 18 |
Hb_000297_020 |
0.2585384639 |
- |
- |
PREDICTED: protein phosphatase 2C 77 [Jatropha curcas] |
| 19 |
Hb_017170_030 |
0.2592474054 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 20 |
Hb_007218_100 |
0.2592734701 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |