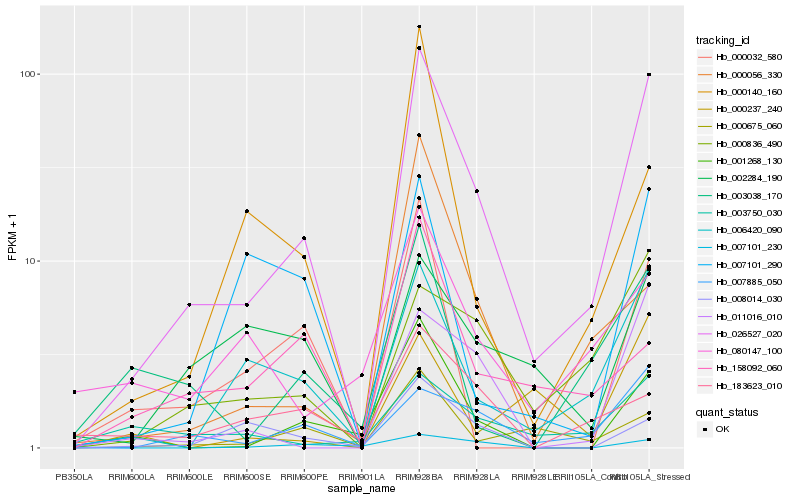
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026527_020 |
0.0 |
- |
- |
PREDICTED: putative methyltransferase DDB_G0268948 [Jatropha curcas] |
| 2 |
Hb_006420_090 |
0.2063236038 |
- |
- |
PREDICTED: zinc finger protein 5 [Jatropha curcas] |
| 3 |
Hb_007885_050 |
0.2203623044 |
- |
- |
PREDICTED: lignin-forming anionic peroxidase-like [Jatropha curcas] |
| 4 |
Hb_001268_130 |
0.2384620966 |
- |
- |
hypothetical protein JCGZ_26837 [Jatropha curcas] |
| 5 |
Hb_011016_010 |
0.2388247073 |
- |
- |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 6 |
Hb_000836_490 |
0.2399937041 |
- |
- |
PREDICTED: E3 SUMO-protein ligase pli1-like [Jatropha curcas] |
| 7 |
Hb_003038_170 |
0.2674214602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000675_060 |
0.269160008 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_000032_580 |
0.272114349 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 10 |
Hb_003750_030 |
0.2774411749 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 11 |
Hb_002284_190 |
0.2810069717 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 12 |
Hb_007101_290 |
0.2847899469 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_008014_030 |
0.2868948189 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 14 |
Hb_080147_100 |
0.294324432 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 15 |
Hb_000140_160 |
0.2956829546 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 16 |
Hb_000056_330 |
0.2966343632 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_183623_010 |
0.2995228555 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 18 |
Hb_007101_230 |
0.2999529524 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_158092_060 |
0.3000867309 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_000237_240 |
0.3013394527 |
- |
- |
- |