| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
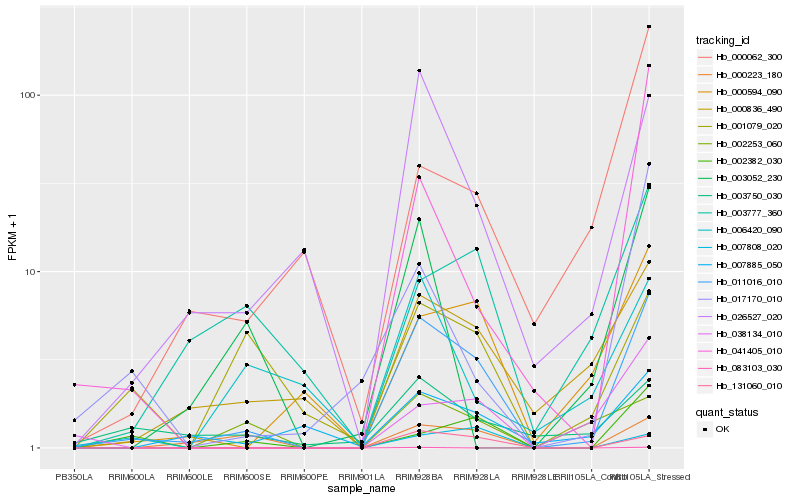
Hb_011016_010 |
0.0 |
- |
- |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 2 |
Hb_131060_010 |
0.2193793249 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 3 |
Hb_026527_020 |
0.2388247073 |
- |
- |
PREDICTED: putative methyltransferase DDB_G0268948 [Jatropha curcas] |
| 4 |
Hb_000836_490 |
0.2612412923 |
- |
- |
PREDICTED: E3 SUMO-protein ligase pli1-like [Jatropha curcas] |
| 5 |
Hb_000223_180 |
0.265657068 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 6 |
Hb_000594_090 |
0.2672457676 |
- |
- |
PREDICTED: allene oxide cyclase 3, chloroplastic-like [Populus euphratica] |
| 7 |
Hb_007885_050 |
0.282167781 |
- |
- |
PREDICTED: lignin-forming anionic peroxidase-like [Jatropha curcas] |
| 8 |
Hb_038134_010 |
0.2948333807 |
- |
- |
- |
| 9 |
Hb_007808_020 |
0.2988437667 |
- |
- |
- |
| 10 |
Hb_002382_030 |
0.3001951315 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Malus domestica] |
| 11 |
Hb_003750_030 |
0.3097609847 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 12 |
Hb_003777_360 |
0.3110776979 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 13 |
Hb_001079_020 |
0.3126498868 |
- |
- |
hypothetical protein POPTR_0012s13720g [Populus trichocarpa] |
| 14 |
Hb_041405_010 |
0.3157390652 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Glycine max] |
| 15 |
Hb_017170_010 |
0.3178158639 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 16 |
Hb_002253_060 |
0.3194359369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006420_090 |
0.321434084 |
- |
- |
PREDICTED: zinc finger protein 5 [Jatropha curcas] |
| 18 |
Hb_000062_300 |
0.3269020529 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 19 |
Hb_003052_230 |
0.3284257969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_083103_030 |
0.3298285407 |
- |
- |
putative retrotransposon protein [Malus domestica] |