| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
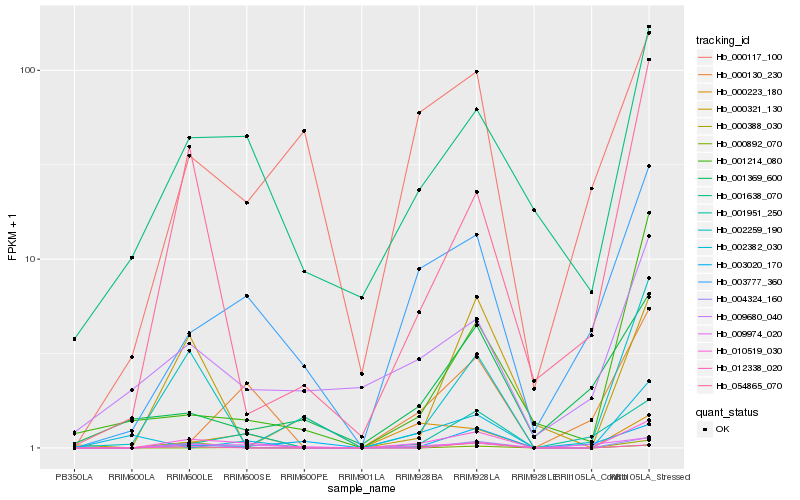
Hb_010519_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_003777_360 |
0.2576261855 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_000223_180 |
0.2709522889 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 4 |
Hb_001638_070 |
0.2731060874 |
transcription factor |
TF Family: bHLH |
Basic helix-loop-helix DNA-binding family protein [Theobroma cacao] |
| 5 |
Hb_004324_160 |
0.2791450565 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 6 |
Hb_001951_250 |
0.281241152 |
- |
- |
- |
| 7 |
Hb_000130_230 |
0.2822864652 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 8 |
Hb_002259_190 |
0.2859124529 |
- |
- |
Omega-3 fatty acid desaturase, endoplasmic reticulum, putative [Ricinus communis] |
| 9 |
Hb_054865_070 |
0.2933527332 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 10 |
Hb_009974_020 |
0.302739899 |
- |
- |
PREDICTED: uncharacterized protein LOC104427753 [Eucalyptus grandis] |
| 11 |
Hb_000321_130 |
0.3152731582 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_009680_040 |
0.315424241 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_000117_100 |
0.3270191857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000892_070 |
0.3285674134 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001214_080 |
0.3310830704 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 16 |
Hb_002382_030 |
0.3358023098 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2-like isoform X2 [Malus domestica] |
| 17 |
Hb_001369_600 |
0.3409841169 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_003020_170 |
0.3518116225 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 19 |
Hb_012338_020 |
0.3532486856 |
- |
- |
- |
| 20 |
Hb_000388_030 |
0.3535783966 |
- |
- |
hypothetical protein JCGZ_13427 [Jatropha curcas] |