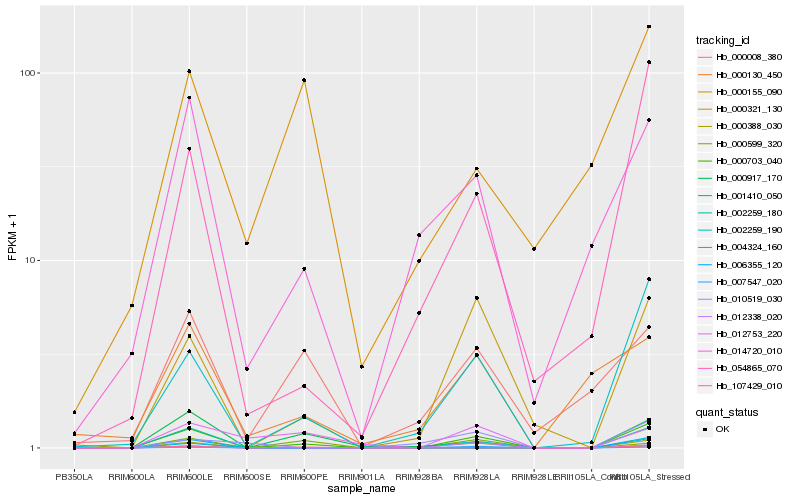
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_040 |
0.0 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 2 |
Hb_002259_190 |
0.1914154075 |
- |
- |
Omega-3 fatty acid desaturase, endoplasmic reticulum, putative [Ricinus communis] |
| 3 |
Hb_004324_160 |
0.222418157 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 4 |
Hb_000321_130 |
0.2292004909 |
- |
- |
Alcohol dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_000917_170 |
0.2563768344 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 6 |
Hb_054865_070 |
0.2658960031 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 7 |
Hb_014720_010 |
0.2993438845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000599_320 |
0.3116584501 |
- |
- |
hypothetical protein JCGZ_17662 [Jatropha curcas] |
| 9 |
Hb_000008_380 |
0.3211972178 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_007547_020 |
0.3285433775 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase A-like [Populus euphratica] |
| 11 |
Hb_012753_220 |
0.3371371067 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18840-like [Populus euphratica] |
| 12 |
Hb_000130_450 |
0.3426982613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_107429_010 |
0.3428633441 |
- |
- |
PREDICTED: calcium-dependent protein kinase 34 [Jatropha curcas] |
| 14 |
Hb_000388_030 |
0.3545238919 |
- |
- |
hypothetical protein JCGZ_13427 [Jatropha curcas] |
| 15 |
Hb_000155_090 |
0.3633433486 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 16 |
Hb_010519_030 |
0.3672536162 |
- |
- |
- |
| 17 |
Hb_001410_050 |
0.369634796 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 18 |
Hb_006355_120 |
0.3700469086 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002259_180 |
0.3720254594 |
- |
- |
- |
| 20 |
Hb_012338_020 |
0.3722695669 |
- |
- |
- |