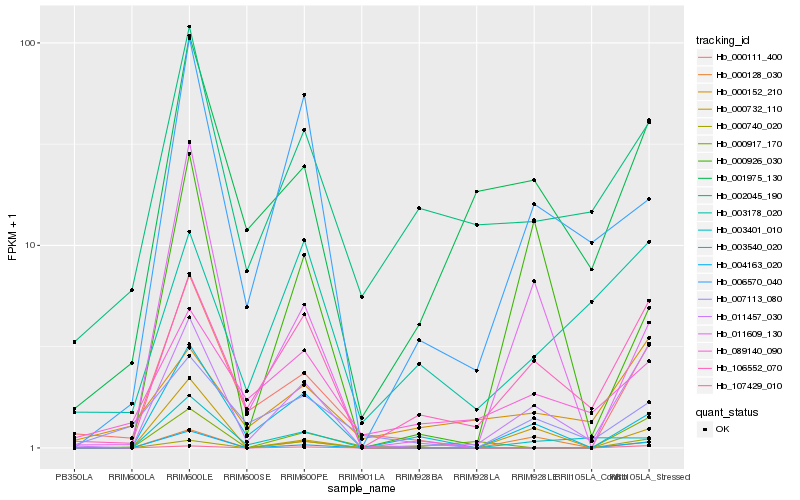
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011457_030 |
0.0 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 2 |
Hb_106552_070 |
0.1676809999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001975_130 |
0.2440542906 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 6 isoform X1 [Populus euphratica] |
| 4 |
Hb_003540_020 |
0.246671954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_107429_010 |
0.2644127483 |
- |
- |
PREDICTED: calcium-dependent protein kinase 34 [Jatropha curcas] |
| 6 |
Hb_000128_030 |
0.2719681375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000732_110 |
0.2745572939 |
- |
- |
hypothetical protein JCGZ_21339 [Jatropha curcas] |
| 8 |
Hb_006570_040 |
0.2756916418 |
- |
- |
hypothetical protein CICLE_v10002683mg [Citrus clementina] |
| 9 |
Hb_004163_020 |
0.2778292276 |
- |
- |
- |
| 10 |
Hb_000152_210 |
0.2853110907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000917_170 |
0.2860756751 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 12 |
Hb_000740_020 |
0.2903983573 |
transcription factor |
TF Family: MYB-related |
asymmetric leaves1 and rough sheath, putative [Ricinus communis] |
| 13 |
Hb_002045_190 |
0.2915994513 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 14 |
Hb_000111_400 |
0.2936706348 |
- |
- |
hypothetical protein POPTR_0003s19310g [Populus trichocarpa] |
| 15 |
Hb_000926_030 |
0.2937704067 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_003401_010 |
0.2948353535 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 17 |
Hb_089140_090 |
0.2975731829 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_011609_130 |
0.2994081416 |
- |
- |
- |
| 19 |
Hb_007113_080 |
0.3010791953 |
- |
- |
PREDICTED: uncharacterized protein LOC105639553 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003178_020 |
0.3049539087 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |