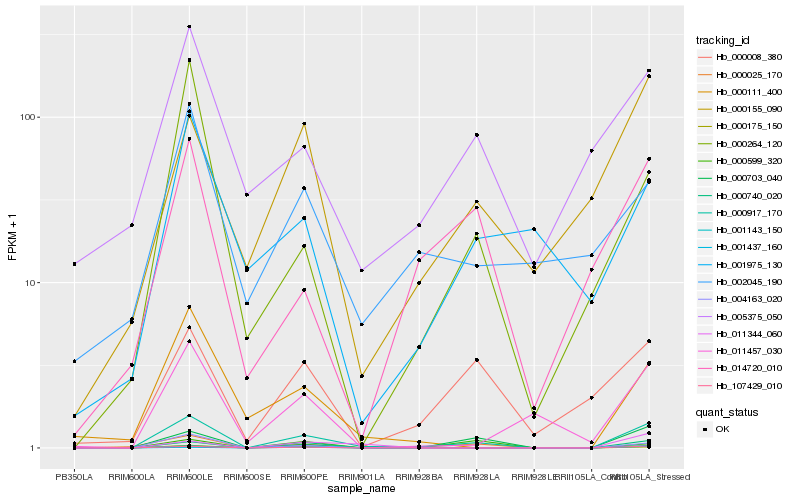
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000917_170 |
0.0 |
- |
- |
PREDICTED: clavaminate synthase-like protein At3g21360 [Jatropha curcas] |
| 2 |
Hb_107429_010 |
0.2295625973 |
- |
- |
PREDICTED: calcium-dependent protein kinase 34 [Jatropha curcas] |
| 3 |
Hb_000703_040 |
0.2563768344 |
- |
- |
hypothetical protein RCOM_1177160 [Ricinus communis] |
| 4 |
Hb_000740_020 |
0.2597022728 |
transcription factor |
TF Family: MYB-related |
asymmetric leaves1 and rough sheath, putative [Ricinus communis] |
| 5 |
Hb_004163_020 |
0.2632214152 |
- |
- |
- |
| 6 |
Hb_000175_150 |
0.2738755977 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein B456_009G311700 [Gossypium raimondii] |
| 7 |
Hb_011457_030 |
0.2860756751 |
- |
- |
type I inositol polyphosphate 5-phosphatase, putative [Ricinus communis] |
| 8 |
Hb_000111_400 |
0.2862070313 |
- |
- |
hypothetical protein POPTR_0003s19310g [Populus trichocarpa] |
| 9 |
Hb_000008_380 |
0.3042582521 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_000264_120 |
0.3059114036 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 11 |
Hb_001437_160 |
0.3116523777 |
- |
- |
PREDICTED: high-affinity nitrate transporter 2.1-like [Jatropha curcas] |
| 12 |
Hb_000155_090 |
0.3126147164 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 13 |
Hb_014720_010 |
0.3159623146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005375_050 |
0.3175124177 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 15 |
Hb_000599_320 |
0.3243325394 |
- |
- |
hypothetical protein JCGZ_17662 [Jatropha curcas] |
| 16 |
Hb_001143_150 |
0.3257473689 |
- |
- |
PREDICTED: mitochondrial protein import protein MAS5-like [Populus euphratica] |
| 17 |
Hb_000025_170 |
0.3280600415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002045_190 |
0.3297068044 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 19 |
Hb_011344_060 |
0.3302188833 |
- |
- |
PREDICTED: uncharacterized protein LOC105645975 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001975_130 |
0.3320432543 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 6 isoform X1 [Populus euphratica] |